Lab
Mosimann Lab
|
|
Statement of Research Interest
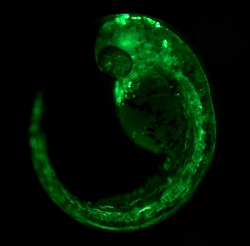
We use zebrafish as our principal model system to study the mechanisms of cell fate determination in development, disease, and evolution. We focus in particular on the emergence, patterning, and cell fate partitioning of the lateral plate mesoderm towards its vast array of downstream cell fates. This includes cardiac, cardiovascular, hematopoietic, kidney, mesothelial and more lineages as also affected in numerous congenital diseases. We have been heavily investing into gene-regulatory element discovery such as enhancers using latest transgenesis approaches, which enables us to make mechanistic discoveries relevant to our understanding of mesoderm evolution, development, and congenital anomalies.
Lab Members
| Moran, Hannah Graduate Student | Kemmler, Cassie L. Technical Staff | Chowdhury, Polina |
