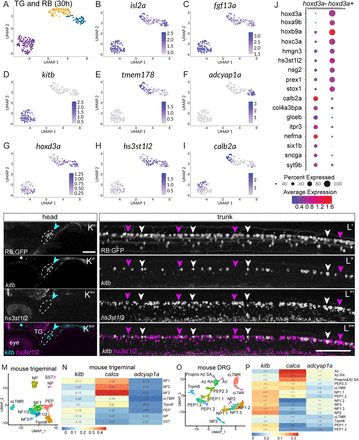
Fig. 3 Comparison of RB and TG transcriptional profiles. A, Unsupervised clustering and UMAP reduction diagram of TG and RB cells derived from 30 hpf TgBAC(neurod1:egfp)nl1 embryos yield three cell clusters containing both TG and RB cells. B,C, Feature plots demonstrating isl2a and fgf13a mark both TG and RB cells. D–F, Feature plots showing presence of RB markers in RB and TG cells. G, Expression of a posterior hox gene, hoxd3a, marks RB cells. H, Feature plot of an RB-specific marker, hs3st1l2. I, Feature plot of a TG-specific marker calb2a. J, Dot plot showing expression of posterior hox genes as well as RB-specific markers (hoxd3a+ subset) and TG-specific markers (hoxd3a− subset). K,L, Lateral views of FISH staining for kitb (magenta arrowheads) and hs3st1l2 (white arrowheads) at 72 hpf in RB (K) or TG (L) neurons marked in RB:GFP larva. Note that subpopulations of RB neurons are positive for kitb or hs3st1l2 while no TG neurons express hs3st1l2. Dashed lines = TG. M, Unsupervised clustering and UMAP reduction diagram of adult mouse TG cells (Yang et al., 2022). N, Heatmap illustrating module scores for each RB cluster that are associated with each type of mouse TG neurons. Note the calca+ RB cluster is most similar to NF3 TG neurons. O, Unsupervised clustering and UMAP reduction diagram of adult mouse DRG cells (Jung et al., 2023). P, Heatmap illustrating module scores for each RB cluster that are associated with each type of mouse DRG neurons. Note the calca+ RB cluster is most similar to Aδ DRG neurons. Scale bar = 100 μm.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ J. Neurosci.