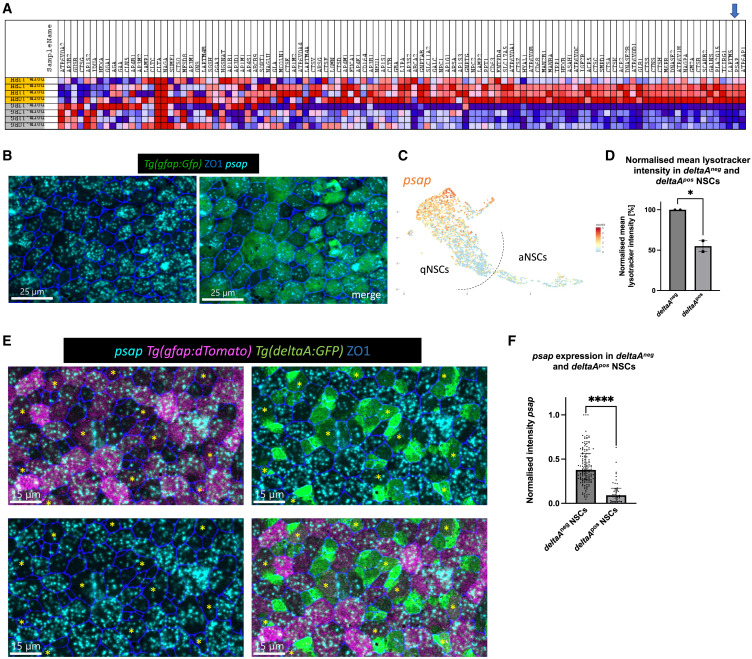
Fig. 2 Psap is heterogeneously expressed in quiescent NSCs (A) Gene expression heatmap of the lysosome KEGG gene set (red = upregulated, blue = downregulated). Each line is 1 biological replicate (gray = aNSCs, yellow = deeply qNSCs – her4high), based on our bulk RNA-seq dataset. Arrow to psap. (B) Whole-mount RNAScope for psap (cyan) and IHC for Tg(gfap:eGFP) (NSCs, green) and ZO1 (tight junctions, blue). Dorsal view of the pallial ventricular zone at 3 MPF. Psap is heterogeneously expressed. Scale bar: 25 μm. (C) Uniform manifold approximation and projection of psap expression in NSCs (scRNA-seq dataset of Morizet et al., 2023). Psap is enriched in cells with deepest/longest quiescence (compare with Figure S4A). (D) Normalized mean LysoTracker intensity in deltaApos and deltaAneg NSCs; bar plot with SD. Each dot is 1 independent experiment with 10 pooled brains. Unpaired t test, ∗p = 0.0218. (E) Whole-mount RNAScope for psap (cyan) and triple IHC for Tg(gfap:dTomato) (magenta), Tg(deltaA:eGFP) (green), and ZO1 (tight junctions, blue). Dorsal view of the pallial ventricular zone at 3 MPF; merged channel, bottom right. Yellow asterisks indicate examples of deltaApos NSCs. Scale bars: 15 μm. (F) Normalized intensity sum of psap after two-dimensional projection in deltaAneg and deltaApos cells within the gfap:dTomatopos NSC population. The highest intensity value has been set to 1. Every dot represents 1 cell; 3 independent experiments. Mann-Whitney test, ∗∗∗∗p ≤ 0.0001.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Stem Cell Reports