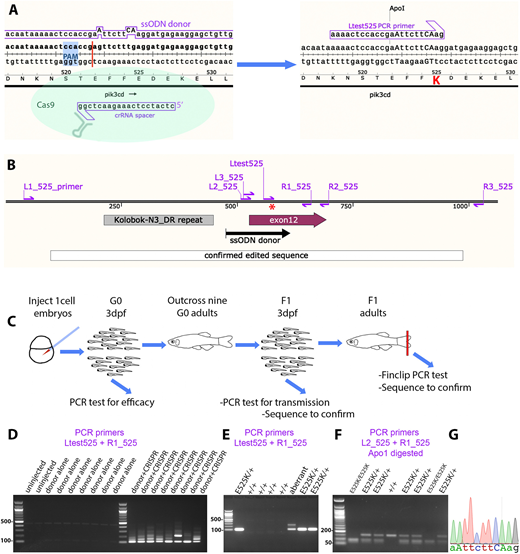
Fig. 1
Homology-directed CRISPR-Cas9 knock-in of a pik3cdE525K allele. (A) Diagram illustrating the knock-in process. The CRISPR-Cas9 cleavage and knock-in is illustrated on the left and the resulting pik3cdE525K allele on the right. On the left, the single-strand oligonucleotide (ssODN) donor sequence and crRNA spacer sequence are positioned alongside the pik3cd DNA sequence and translated amino acid sequence, with the CRISPR-Cas9 protospacer adjacent motif (PAM) highlighted in blue and the cleavage site shown as a red line. On the right, mutated DNA bases are shown in upper case, the E525K coding change is highlighted in red, and the introduced ApoI cleavage site is indicated. The Ltest525 PCR primer is specific for the knock-in allele. (B) Map of the zebrafish pik3cdE525K locus. PCR primers are shown as magenta arrows; the mutation site is marked with a red asterisk; and positions are shown for the ssODN donor used for knock-in, the region confirmed by sequencing after knock-in, pik3cd exon 12 and a DNA repeat incompatible with PCR primer location. (C) Diagram illustrating the workflow for establishing the line with the pik3cdE525K allele. (D) Agarose electrophoresis gel with knock-in-specific test PCRs from 3 days post fertilisation (dpf) G0 embryos. All the embryos previously injected with CRISPR-Cas9 and ssODN donor produced the expected 103 bp amplicon and also lower electrophoretic mobility products. PCRs from uninjected embryos, or embryos injected with CRISPR-Cas9 alone, did not amplify. DNA size marker is a 100 bp ladder. (E) Agarose electrophoresis gel with knock-in-specific test PCRs from 3 dpf F1 embryos from an outcross of a G0 adult that had been injected with CRISPR-Cas9 and ssODN donor when an embryo. PCRs from embryos with the pik3cdE525K allele produced a 103 bp amplicon. A lower-mobility band indicates a putative aberrant knock-in event. DNA size marker is 100 bp ladder. (F) Agarose electrophoresis gel with Apo1-digested PCR amplicons from F2 embryos from a pik3cdE525K/+ in-cross. The pik3cdE52K5 allele introduces an Apo1 site, allowing pik3cdE525K/E525K, pik3cdE525K/+ and pik3cd+/+ genotypes to be distinguished. DNA size marker is 50 bp ladder. (G) Sanger sequencing trace from a PCR amplicon across the knock-in locus of a pik3cdE525K/E525K F2 adult. The mutated DNA bases are shown in upper case as in A.