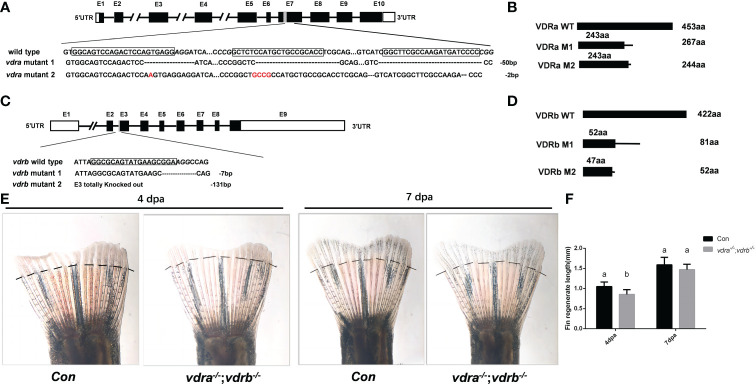
Figure 1
Generation of zebrafish vdra- and vdrb –deficient using CRISPR/Cas9 technique. (A) Targeted depletion of the vdra gene; The CRISPR/Cas9 target site was located in exon 7. Two genotypes, 50-bp deletion and 2-bp deletion, were used to establish the vdra knockout line (highlighted in red); (B) The diagram shows the predicted VDRa protein from wild-type, M1, and M2 zebrafish. M1 consisted of 243 amino acids identical to WT (green) and 24 miscoding amino acids (black); M2 consisted of 243 amino acids identical to WT and one miscoding amino acid (black); (C) Targeted depletion of the vdrb gene; The CRISPR/Cas9 target site was located in exon 3. Two genotypes, 7-bp deletion and 131-bp deletion, were used to establish the vdrb knockout line (highlighted in red); (D). The diagram shows the predicted VDRb protein from wild-type, M1, and M2 zebrafish. M1 consisted of 52 amino acids identical to WT (green) and 29 miscoding amino acids (black); M2 consisted of only 47 amino acids identical to WT and five miscoding amino acids (black). (E) Zebrafish(100dpf) caudal fin regeneration at 4 and 7days post amputation. Dashed lines indicated approximate amputation planes. (F). Statistical analysis of the regeneration length. Six fish were used in each group. The bars with different letters indicated significant difference(p<0.05).