Fig. 1
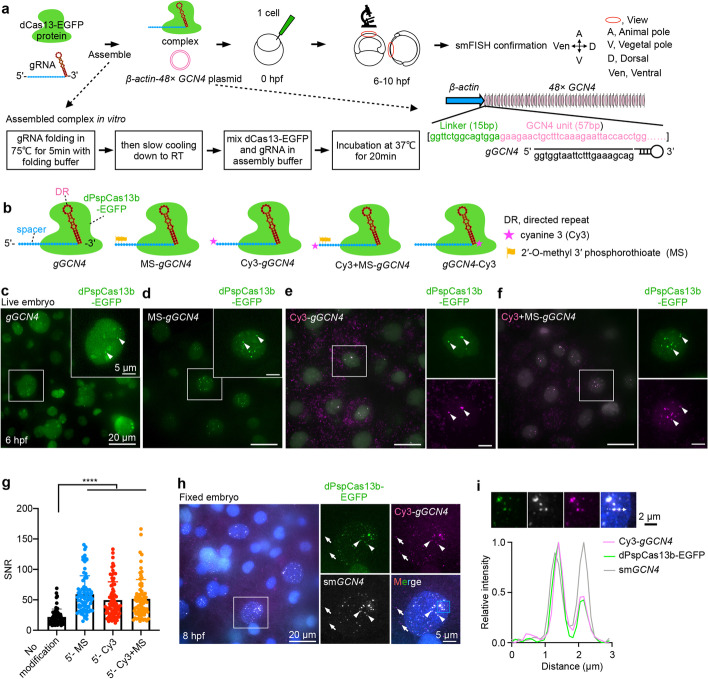
Imaging ectopic GCN4 RNA repeats by CRISPR-dCas13 in zebrafish embryos. a A schematic screen to identify robust CRISPR-dCas13 systems to visualize RNAs in zebrafish embryos. 5.6 μM dCas13-EGFP protein assembled with 8.4 μM in vitro transcribed gRNA (gGCN4) at a molar ratio 1:1.5, plus 5.9 nM (50 ng/μL) β-actin-48× GCN4 plasmid were injected into zebrafish embryos at the 1-cell stage. For some lower stability dCas13 constructs, we increased the concentration (see “Methods”). After injection, fluorescent signals in the nucleus were detected at 6–10 hpf (hours post fertilization). The CRISPR-dCas13 detected signals were confirmed by smFISH. β-actin-48× GCN4, zebrafish β-actin promoter derives 48× GCN4 expression. b A schematic view of chemically modified gRNA. Cyanine 3 (Cy3) and 2′-O-methyl 3′ phosphorothioate (MS) modifications at the 5′ and/or 3′ end of gGCN4 were used, including MS-gGCN4, Cy3-gGCN4, Cy3+MS-gGCN4 and gGCN4-Cy3. The gRNA of dCas13b consists of a 5′ spacer sequence (blue region) and a 3′ direct repeat sequence (DR, red region) [43]. c–f Chemically modified gRNA improves CRISPR-dCas13 in RNA labeling. Representative images of CRISPR-dPspCas13b system-labeled 48× GCN4 with gGCN4 (no modification, c), MS-gGCN4 (d), Cy3-gGCN4 (e), and Cy3+MS-gGCN4 (f) at 6 hpf in live embryos, respectively. White arrowheads indicate labeled signals (c–f) and colocalization signals between Cy3 and EGFP (e, f). g SNR statistics of the 48× GCN4 signals labeled by unmodified and chemically modified of gGCN4 with dPspCas13b-EGFP. Data from about 10 embryos in each group, n = 71, 80, 73, 74 cells. h smFISH confirms the CRISPR-dPspCas13b system targeting 48× GCN4 at 8 hpf in fixed embryos. White arrowheads indicate colocalization of GCN4 smFISH, Cy3-gGCN4, and dPspCas13b-EGFP. White arrows indicate non-specific signals. Blue box, zoomed in i. i Analysis of the blue box region in h. Line scan of the relative fluorescence intensity of signals (white dot line arrow in upper panel) shows the colocalization of Cy3-gGCN4, dPspCas13b-EGFP, and smGCN4 FISH. Scale bar 2 μm. In c-f and h, data scale bar 20 μm; the white box indicates magnified area, scale bar 5 μm. In g, data is represented as mean ± SD, unpaired two-tail Student’s t test, **** p< 0.0001