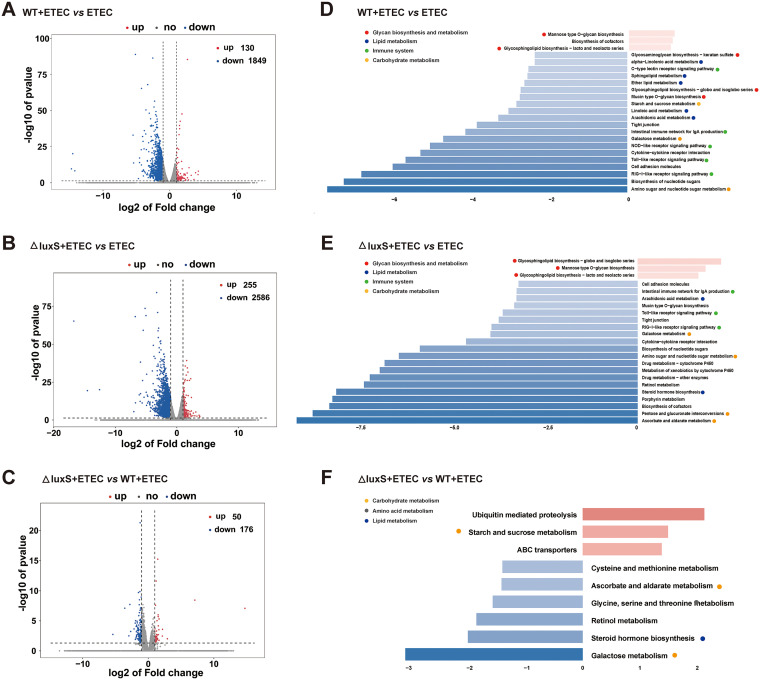
FIG 5
Transcriptomic analysis of zebrafish larvae. (A–C) Volcano plots of DEGs. Log2 (Fold change) ≥1 was set as the threshold. (D–F) Pathway classification based on KEGG enrichment analysis of DEGs. We enriched the differential genes according to Log2 (Fold change) ≥1, upregulated genes