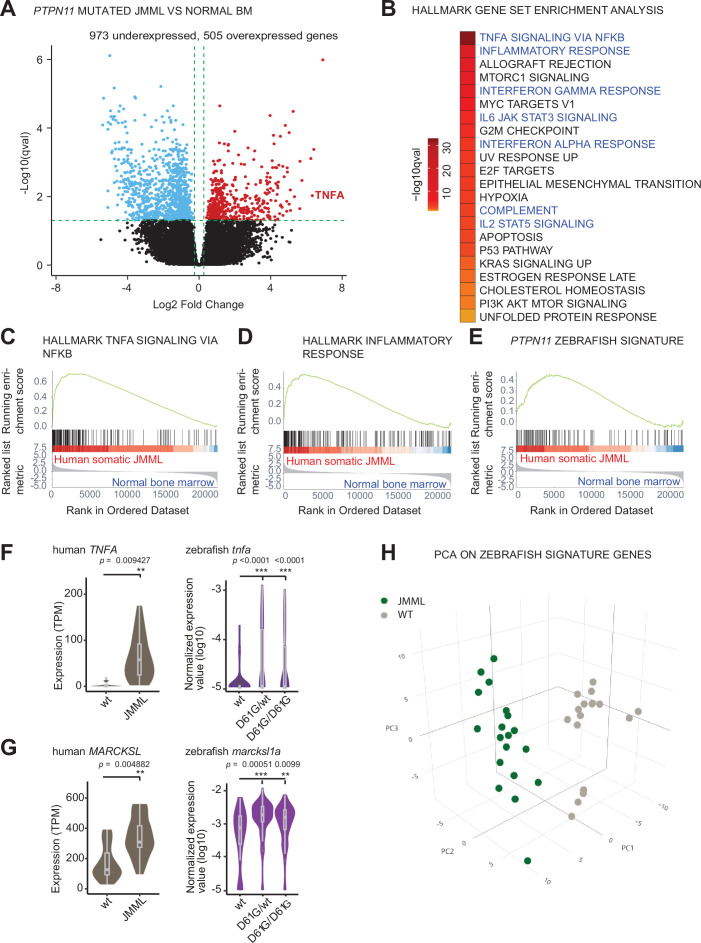
Figure 5.
(A) Volcano plot of differentially expressed genes of HSPCs derived from bone marrow of JMML patients with PTPN11 mutations (n = 5) and healthy bone marrow (n = 7). Underexpressed genes are marked in blue and overexpressed genes are marked in red. TNFA expression is highlighted. Green dashed lines indicate the significance level. (B) Gene set enrichment analysis (GSEA) for the MSigDB’s hallmark gene sets in HSPCs from JMML compared to normal human age-matched bone marrow. GSEA plots for TNFA_SIGNALING_VIA_NFKB (C), INFLAMMATORY RESPONSE (D), and the custom zebrafish signature based on the top 100 human orthologous of genes upregulated in monocyte/macrophage progenitor cluster of zebrafish HSPCs (E). Violin plots show the expression of TNFA (F) and MARCKSL (G) in either human wild type (wt) vs. JMML HSPCs, and zebrafish Shp2wt vs. Shp2D61G/wt and Shp2D61G/D61G monocyte/macrophage HSPC progenitors. **p < 0.01, ***p < 0.001, t-test. (H) Principal component analysis (PCA) for the 100 genes included in the custom zebrafish signature. Green dots represent PTPN11 mutated JMML, gray ones represent normal human age-matched bone marrow. PC1: 19% of the variance, PC2: 16% of the variance, PC3: 14% of the variance.
Similar molecular signatures in hematopoietic stem and progenitor cells (HSPCs) from human juvenile myelomonocytic leukemia (JMML) patients and zebrafish Shp2D61G embryos.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Elife