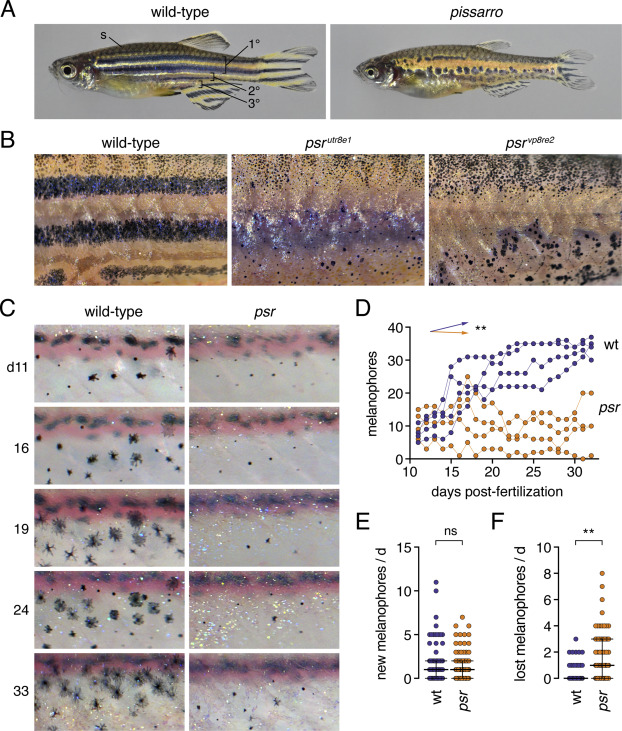
Fig. 1 Phenotypes and pattern ontogeny of wild-type and pissarro (psr) mutant zebrafish. (A) Wild-type adult fish with primary, secondary and tertiary pattern elements and scale (s) melanophores labeled. psr mutants (here psrvp8re2) have disrupted melanophore stripes and fins that are short relative to body size. (B) Higher magnification images illustrating disorganized melanophores and iridophores in wild-type fish or homozygous mutants for different psr alleles. In psrvpr8e2 some melanophores were apparent on ventral scales (arrowhead), typical of the wik genetic background in which the allele was maintained. Insets illustrate well-spread appearance of most stripe melanophores in wild-type and heterogeneity of melanophore morphologies in psr mutant alleles. (C) Repeated daily imaging of representative wild-type and psrutr8e1 mutant larvae through adult pigment pattern formation. Days at left indicate days post-fertilization. Boxes indicate newly differentiating melanophores. Colored circles indicate melanophores lost from the pattern. These same fish are shown in Movies S1, S2. (D) Total numbers of melanophores increased in each of 4 repeatedly imaged wild-type larvae but remained at initial levels or decreased in each of 4 psr mutants in ventral portions of 3 segments. Differences in melanophore numbers were reflected in a significant genotype × day interaction after controlling for variation among individuals (∗∗, F1,6 = 23.7, P = 0.0028; arrows indicate slopes of partial regression coefficients). (E) Numbers of newly differentiating melanophores per day did not differ between wild-type and psr mutants after controlling for variation among individuals (ns, F1,6 = 0.3, P = 0.6099). (F) Melanophores were, however, significantly more likely to be lost from psr mutants than wild-type after controlling for variation among individual fish (∗∗, F1,6.2 = 16.5, P = 0.0063; counts of melanophores were square root-transformed for analysis to correct for heterogeneity in residual variance between genotypes). Bars in E and F are medians ± interquartile range.
Reprinted from Developmental Biology, 476, Eom, D.S., Patterson, L.B., Bostic, R.R., Parichy, D.M., Immunoglobulin superfamily receptor junctional adhesion molecule 3 (Jam3) requirement for melanophore survival and patterning during formation of zebrafish stripes, 314-327, Copyright (2021) with permission from Elsevier. Full text @ Dev. Biol.