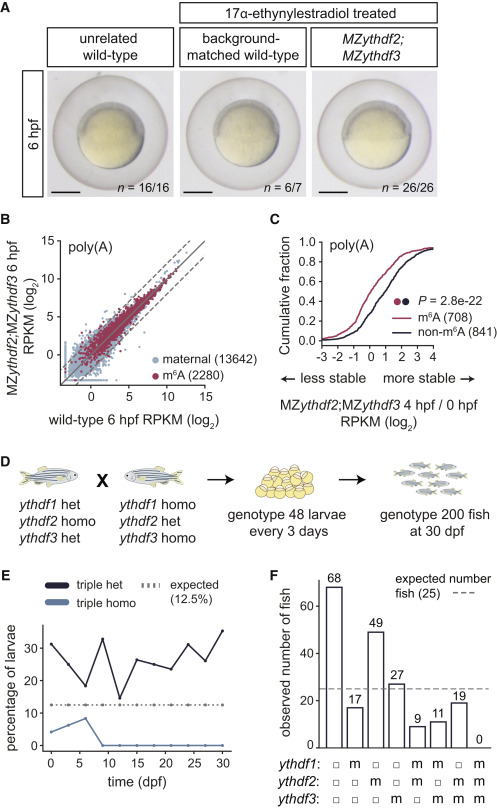
Fig. 5 Figure 5. Triple Loss of Ythdf Readers Disrupts Zebrafish Development (A) MZythdf2;MZythdf3, background-matched wild-type, and unrelated TU-AB wild-type zebrafish embryos develop at similar rates. Parents of mutant and background-matched control embryos were 17α-ethynylestradiol treated. n, replicate number of embryos at same developmental stage. Scale bars, 500 μm. (B) Biplot of expression (log2 RPKM) of maternal (n = 13,642) and m6A-modified (n = 2,280) mRNAs between wild-type and MZythdf2;MZythdf3, from 6 hpf poly(A) mRNA-seq. Dashed lines, 2-fold change. (C) Cumulative distribution of fold changes in maternal mRNA abundance (log2 RPKM) between 4 and 0 hpf in MZythdf2;MZythdf3 embryos, for m6A-modified (n = 708) and non-modified (n = 841) mRNAs, from poly(A) mRNA-seq. p values computed by a Mann-Whitney U test. (D) Schematic of cross and genotyping strategy for triple Ythdf mutants. Female fish (ythdf1+/−;ythdf2−/−;ythdf3+/−) were crossed to males (ythdf1−/−;ythdf-+/−;ythdf3−/−) to generate triple homozygotes (1 of 8 possible genotypes). Every 3 days, 48 larvae were genotyped, with 200 more fish genotyped at 30 dpf. (E) Percentage of triple heterozygous (het) or triple homozygous (homo) fish during development. Dotted line, expected percentage (12.5%) of each genotype, from cross in (D). (F) Number of fish with each genotype from cross in (D) at 30 dpf. For each ythdf allele: filled circle, heterozygous; m, homozygous. Dotted line, expected fish number (25), equal for all genotypes.
Image
Figure Caption
Figure Data
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Cell Rep.