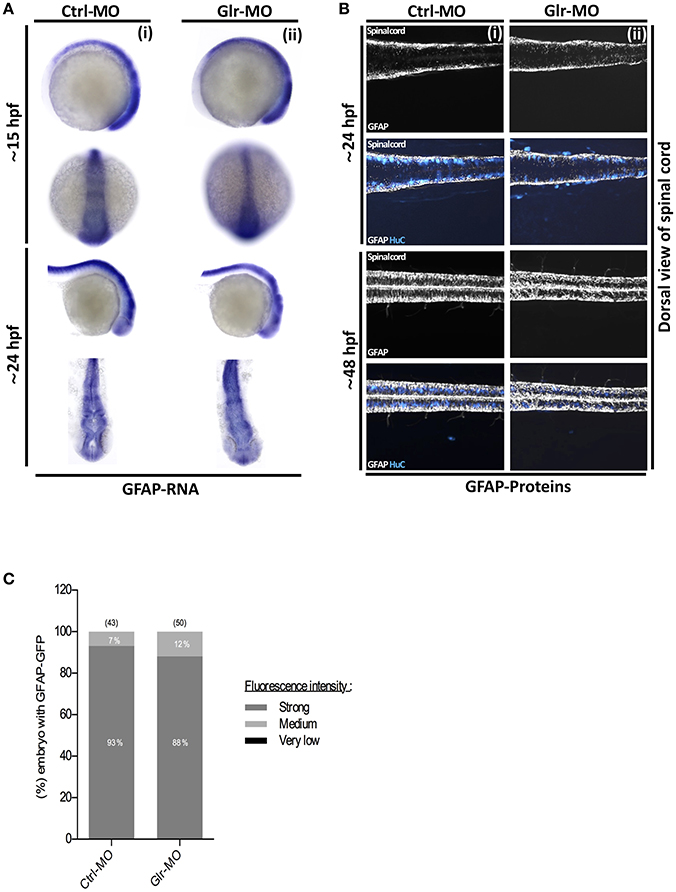
Fig. 3
Glycine signaling defect does not affect GFAP+ NSCs subpopulation. (A) Using GFAP as a marker of NSCs subpopulation, GFAP transcripts were analyzed by whole-mount in situ hybridization. No major difference of GFAP+ subpopulation was observed between Ctrl-MO (i), and Glr-MO (ii) conditions at 15 and 24 hpf. (B) Double tg(GFAP:GFP; HuC:RFP) line was used to analysis GFAP expression (white) and HuC+ expression as mature neurons marker (Bleu). GFAP+ signal does not change between Ctrl-MO (i) and Glr-MO (ii) conditions at 24 and 48 hpf. However, a clear decreasing of HuC+ signal was observed between Ctrl-MO (i), and Glr-MO (ii) conditions. (C) Quantification of GFAP-GFP embryos upon disruption of glycine signaling does not affect GFAP+ subpopulation. For both ISH and IHC, n > 20 embryos per sample. IHC, immunohistochemistry; ISH, in situ hybridization.