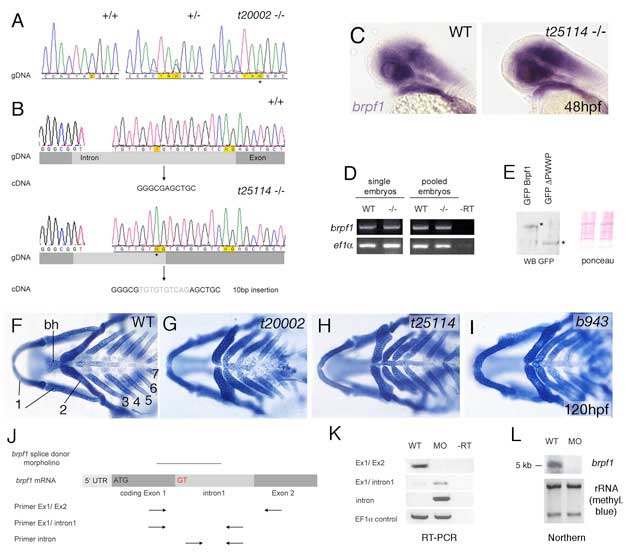
Fig. S1 Analysis of bpfr1 mutations and antisense MO. (A) Sequencing profiles of brpf1 genomic fragment from wild-type (+/+), heterozygous (+/-) and t20002 mutant (-/-) animals. In the mutant allele, TAC is mutated to a TAA stop codon, as indicated by an asterisk. (B) Sequencing profiles of brpf1 genomic fragment from wild-type (+/+, upper panel) and t25114 mutant animals (-/-, lower panel). The t25114 mutation generates a new splice acceptor site within the intron (AG), which is preferentially used. This leads to a 10 bp insertion (indicated in gray) and a frameshift in the cDNA. (C-E) The t25114 mutant brpf1 transcript and the resulting C-terminally truncated Brpf1 protein are stable. (C) Whole-mount in situ hybridization with brpf1 probe of wild-type (left) and t25114 homozygous embryo at 48 hpf. Embryos were genotyped after photography. (D) Semi-quantitative RT-PCR to amplify brpf1 (upper panels) or, as control, ef1α (lower panels) fragments from cDNA of single (left) or pooled (right) wild-type (WT) or t25114 homozygous larvae (-/-) at 120 hpf, when the phenotype was morphologically visible. Mutant bands were cloned, over 50 clones were sequenced and all found to contain the 10 bp insertion (data not shown). PCR primers used were: brpf1-F, TTCTTCACTGAGCCCGTACC and -R, GGGGACCAGAGACTTTAGGG; ef1α-F, TCACCCTGGGAGTGAAACAGC and -R, ACTTGCAGGCGATGTGAGCAG. (E) Anti-GPF western blot analysis of protein extracts from zebrafish embryos (10 hpf) after injection of plasmid DNA encoding GFP fusion proteins of full-length Brpf1 or C-terminally truncated Brpf1 lacking the PWWP domain (left). Full-length and truncated proteins are present in comparable amounts. (Right panel) Ponceau Red staining of nitrocellulose filter as loading control. (F-I) The three mutant brpf1 alleles display skeletal defects of similar strengths. Panels show Alcian Blue staining of heads at 120 hpf, ventral views. Numbers of pharyngeal arches and basihyal (bh) of second arch are indicated in F. Note that the t25114 allele (H), which only lacks the C-terminal PWWP domain, completely lacks the basihyal and the hypobranchials of arches 3 and 4, similar to the potential null allele t20002 (G; compare with Fig. 1G). (J-L) The brpf1 splice donor MO efficiently blocks splicing of intron 1 in vivo. (J) Diagram demonstrating of the structure of unspliced brpf1 hnRNA and the positions of the MO and the primers used in K. Primer sequences are available upon request. (K) RT-PCR on cDNA from uninjected and brpf1 morphant larvae at 48 hpf, using the primer pairs indicated in J. The wild-type cDNA (first lanes) only gave a band of appropriate size with a primer pair from exons 1 and 2 (panel 1), but no band with a primer pair from exon 1 and intron 1 (panel 2), or with two intron-internal primers (panel 3), whereas the morphant cDNA (second lanes) yielded the opposite pattern, indicating that intron 1 was not spliced out. Lanes 3 contain -RT controls, ruling out contamination with genomic DNA. Row 4 shows ef1α loading controls. (L) Northern blot analysis with brpf1 probe of total RNA from wild-type (lane 1) or morphant (lane 2) embryos. Only the wild-type RNA shows a brpf1 band of the appropriate size (5 kb). No band was detected in the morphant RNA, indicating that most of the unspliced brpf1 transcripts are degraded. (Lower panel) Methylene blue staining of blot showing rRNA as loading control.
Image
Figure Caption
Figure Data
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Development