Fig. S5
Appl mutants
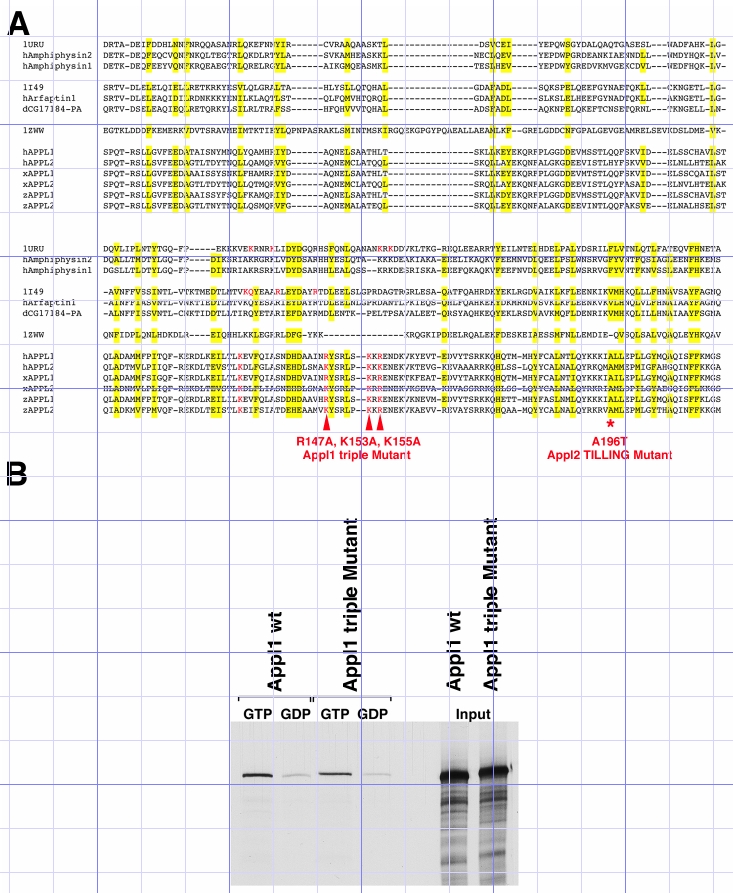
(A) Multiple Sequence Alignment of the BAR domain from members of the Amphiphysin-, Arfaptin-, Endophilin- and APPL- families. Conserved residues between all family members are highlighted in yellow. Residues required for membrane interaction of Amphiphysin (1URU) and Arfaptin (1I49) (Peter et al., 2004) as well as corresponding residues in APPLs are shown in red. Three of those residues in Appl1 were mutated to Alanine (R147, K153, K155; corresponding to residues 146, 152, 154 in the human protein) and are referred to as Appl1 triple Mutant.
A conserved Alanine (A196) in Appl2 was hit by tilling (Appl2 tilling mutant). Abbreviations and Accession numbers are as follows: h: Homo sapiens; d: Drosophila melanogaster; x: Xenopus laevis; z: Danio rerio; hAmphiphysin: NP_647594; hAmphphysin2: NP_647593; hArfaptin1: NP_001020766; dCG17184-PA: NP_650058; hAPPL1: NP_036228; hAPPL2: NP_060641; xAPPL1: NP_001083547; xAPPL2: NP_001087772; zAPPL1 (for Appl1): EU053152; zAPPL2 (for Appl2): EU053153.
(B) GST-pull down assays using recombinant Rab5C protein, loaded with either GTPγS (+GTP) or GDP (+GDP) nucleotides. In vitro translated Appl1 wt and Appl1 triple Mutant proteins are bound specifically by the activated, GTP-bound form of the small GTPase.
Reprinted from Cell, 133(3), Schenck, A., Goto-Silva, L., Collinet, C., Rhinn, M., Giner, A., Habermann, B., Brand, M., and Zerial, M., The endosomal protein Appl1 mediates Akt substrate specificity and cell survival in vertebrate development, 486-497, Copyright (2008) with permission from Elsevier. Full text @ Cell