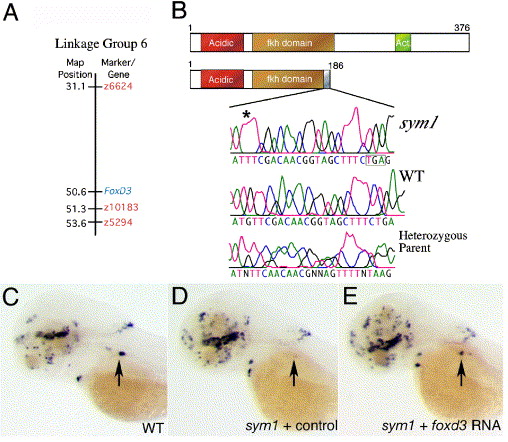
Fig. 4 sym1 is an inactivating mutation in foxd3. (A) Linkage of the foxd3 gene to the CA repeat markers on zebrafish chromosome 6. The positions of markers z10183, z6624 and z5294, are highlighted in orange (source http://zfin.org), while foxd3 is highlighted in blue. The z10183 marker was mapped to within 1 cM of the foxd3 gene. (B) Schematic diagrams of the wild-type and mutant Foxd3 proteins. In sym1, a guanine (537) deletion results in a frame shift leading to a premature stop codon and disruption of the encoded DNA-binding domain. An acidic domain is highlighted in orange at the N-terminus, and the C-terminus transcriptional effector domain is shown in green. Chromatogram traces below the schematic illustrate the nucleotide change (asterisk) affecting the foxd3-coding region. (C–E) Injection of foxd3 RNA rescues the sympathetic th-expression pattern in sym1 mutant embryos. Dorso-lateral views at 4 dpf of wild-type (C) and sym1 mutant embryos (D, E) labeled with the th riboprobe. Wild-type siblings contain 10–40 th-expressing cells in the cervical complex at this stage (C), whereas the sym1 mutant embryos injected with control RNA have 0–2 th-expressing cells (D). Injection of full-length foxd3 RNA into sym1 mutant embryos results in rescue (E), defined as >10 th-expressing sympathetic neurons in the region of the cervical complex. Arrows indicate position of the cervical complex.
Reprinted from Developmental Biology, 292(1), Stewart, R.A., Arduini, B.L., Berghmans, S., George, R.E., Kanki, J.P., Henion, P.D., and Look, A.T., Zebrafish foxd3 is selectively required for neural crest specification, migration and survival, 174-188, Copyright (2006) with permission from Elsevier. Full text @ Dev. Biol.