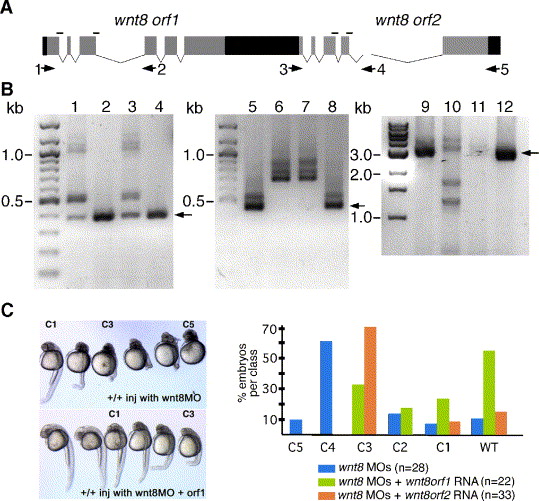
Fig. S1 Characterization of wnt8 splice blocking MOs. (A) Structure of the wnt8 locus. Boxes denote exons, black shading represents non-coding regions. wnt8 orf1 and orf2 are the two wnt8 coding regions. Black lines above exons indicate positions of splice blocking MO sequences. Arrows indicate primers used for RT-PCR characterization. (B) RT-PCR analysis of wnt8 splice blocking efficiency at shield stage. Lanes 1–4: PCR using primers 1 and 2. Lanes 5–8: PCR using primers 3 and 4. Lanes 9–12: PCR using primers 1 and 5. Lane 1, 5, 9: RNA was prepared from ∼ 80 embryos injected with orf1 splice blocking MOs. Lanes 2, 6, 10: RNA was prepared from ~ 80 embryos injected with orf2 splice blocking MOs. Lanes 3, 7, 11: RNA was prepared from ∼ 80 embryos injected with all MOs. Lanes 4, 8, 12: RNA was prepared from wild type embryos. Molecular weight standard sizes are indicated (kb). Arrows indicate the size of the expected wild-type products. Note that the orf1 splice blocking MOs prevent complete splicing of the orf1 transcripts (lane 1) but not orf2 transcripts (lane 2) and orf2 splice blocking MOs do not block orf1 splicing (lane 5) but efficiently block orf2 splicing (lane 6). Note the almost complete absence of wild-type full-length transcripts when four splice blocking MOs are injected (lane 11). (C) Rescue of the splice-blocker-induced phenotype by wild-type RNA. Since the splice blocking MOs bind to unspliced exon/intron junctions, wild-type RNA is not affected by the MOs. Left: examples of phenotypes observed in wild-type embryos injected with wnt8 MOs or in embryos injected with both wnt8 MOs and wild-type wnt8 orf1 RNA. C1–C5 are dorsalization classes described by Mullins et al. (1996) where C5 is the strongest dorsalized phenotype and C1 is characterized by the loss of the ventral tail fin. Right: graph of rescue experiment results. In this experiment, wnt8 RNA (concentration = 10 ng/μl) was injected into embryos previously injected with MOs. RT-PCR Methods: approximately 80 embryos from each treatment were homogenized in Trizol (Invitrogen) and processed according to the manufacturer′s instructions. 0.5–1.0 μg RNA was used for first strand cDNA synthesis with Thermoscript primed with an oligo-dT primer (Invitrogen). Forty PCR cycles were performed for the results shown in this figure.
Reprinted from Developmental Biology, 287(2), Ramel, M.C., Buckles, G.R., Baker, K.D., and Lekven, A.C., WNT8 and BMP2B co-regulate non-axial mesoderm patterning during zebrafish gastrulation, 237-248, Copyright (2005) with permission from Elsevier. Full text @ Dev. Biol.