Fig. 2
- ID
- ZDB-FIG-210514-22
- Publication
- Vona et al., 2021 - A biallelic variant in CLRN2 causes non-syndromic hearing loss in humans
- Other Figures
- All Figure Page
- Back to All Figure Page
|
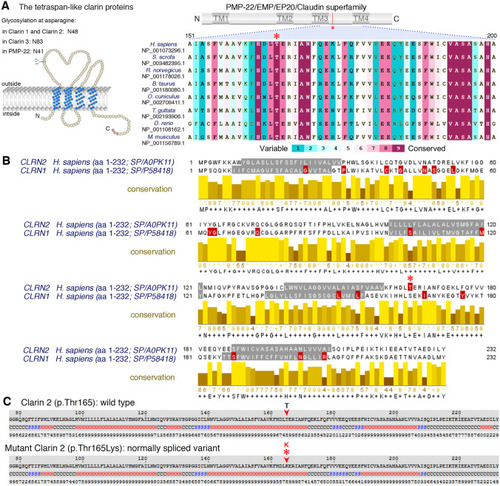
Conservation of the p.Thr165 residue, and clarin 1/clarin 2 alignment. |