Fig. S5
- ID
- ZDB-FIG-170609-11
- Publication
- Thyme et al., 2016 - Polq-Mediated End Joining Is Essential for Surviving DNA Double-Strand Breaks during Early Zebrafish Development
- Other Figures
- All Figure Page
- Back to All Figure Page
|
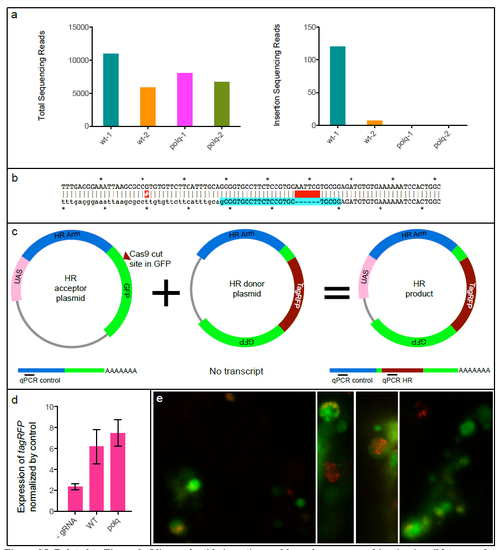
Related to Figure 3. Oligonucleotide insertion and homologous recombination in wild type and MZpolq embryos. a) Total number of sequencing reads and number of reads containing an insertion for oligonucleotide-injected wild-type (wt) and MZpolq mutant embryos. b) Sequence of the insertion. The resulting allele (top) is an in-frame insertion of a six base-pair EcoRI site (GAATTC) at the camk2g locus (bottom). No basepairs were lost or gained outside of the insertion in this experiment. A single-nucleotide polymorphism between the zebrafish used and reference sequence is annoted with a red-highlighted #. c) Schematic of assay used to quantitatively assess homologous recombination. Homologous recombination generates fusion gene of UAS and tagRFP. Injection of mRNA encoding GAL4 generates GAL4 protein, which binds to UAS and activates transcription. Transcripts are detected by qRT-PCR. Approximate locations of qPCR primers are shown for the transcripts produced from each plasmid. d) Expression of tagRFP mRNA normalized to expression of homology arm RNA. Homologous recombination is stimulated by Cas9-induced DSBs in both MZpolq mutant and wild-type embryos. e) Example images of MZpolq mutant cells in which homologous recombination events occurred as detected by tagRFP fluorescence. |