Fig. 1
- ID
- ZDB-FIG-160913-1
- Publication
- Veldman et al., 2015 - The N17 domain mitigates nuclear toxicity in a novel zebrafish Huntington's disease model
- Other Figures
- All Figure Page
- Back to All Figure Page
|
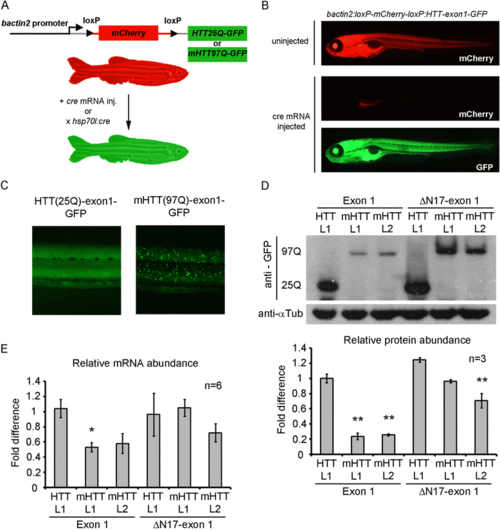
Design and demonstration of Cre-loxP inducible HTT-exon 1-GFP expression in transgenic zebrafish. a Schematic of the transgene design using the bactin2 promoter to drive expression of floxed mCherry followed by the HTT-exon 1 cassette being tested. Following Cre expression by mRNA injection or HS:cre transgene induction, mCherry is recombined out and the desired HTT-exon 1 cassette fused to GFP is expressed ubiquitously. b Photo micrographs depicting 5 day old, HTT-exon1(25Q) embryos displaying mCherry expression when Cre is absent (upper panel) or mCherry negative, HTT-exon1-GFP positive expression following cre mRNA injection (bottom two panels). c Close-up view of the trunk of HTT(25Q)-exon1-GFP and mHTT(97Q)-exon1-GFP embryos demonstrating ubiquitous, diffuse expression of HTT-exon1-GFP (left panel) and ubiquitous, aggregated expression of mHTT-exon1-GFP (right panel). d A representative western blot and densitometric measurments for each transgenic line (n = 3). Anti-GFP antibody (top panel) and anti-tubulin loading controls (bottom panel) on protein isolated from pools of 5 day old embryos for each HTT transgenic line crossed to HS:cre. 97Q and 25Q mark the location of mHTT-GFP and HTT-GFP protein bands respectively. Protein expression was highest in HTT-exon1 L1 and HTT-ΔN17-exon1 L1 lines. mHTT-exon1 line 1(L1) and line 2(L2) both express at similar levels as do mHTT-ΔN17-exon1 line 1(L1) and line 2(L2). Densitometric measurements (lower bar graph) demonstrate significantly lower levels of protein in mHTT-exon1 L1 and L2 compared to all other lines while mHTT-ΔN17-exon1 L2 is significantly lower than HTT-exon1 L1 and HTT-ΔN17-exon1 L1. Comparisons were done by one-way ANOVA with Bonferroni posthoc test, **p < 0.01, error bars are SEM. e Quantification of transgene expression by quantitative RT-PCR. Six pools of ten embryos from separate clutches of each HTT transgenic line crossed to HS:cre were analyzed. Comparisons of expression level were done by one-way ANOVA with Bonferonni posthoc test, *p < 0.05, error bars are SEM |