Fig. 1
- ID
- ZDB-FIG-160229-11
- Publication
- Cao et al., 2016 - Single epicardial cell transcriptome sequencing identifies Caveolin-1 as an essential factor in zebrafish heart regeneration
- Other Figures
- All Figure Page
- Back to All Figure Page
|
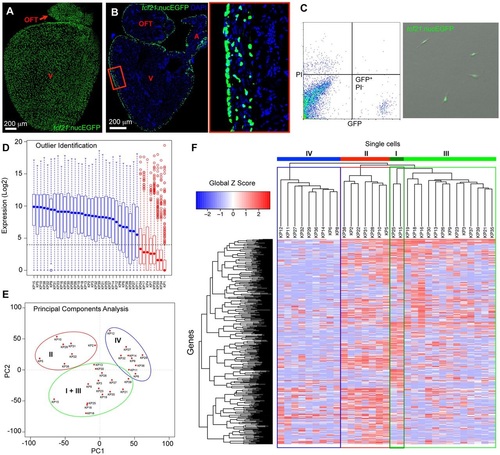
Single-cell transcriptome sequencing of adult zebrafish epicardial cells. (A,B) Whole-mount (A) and section (B) images of an adult tcf21:nucEGFP heart. The boxed region is enlarged to the right. OFT, outflow tract; A, atrium; V, ventricle. Blue, DAPI. (C) FACS isolation of GFP-positive, propidium iodide (PI)-negative, epicardial cells from uninjured hearts of tcf21:nucEGFP adult fish. Drops of isolated cells were plated into a culture dish with DMEM plus 10% FBS for 24h, to examine survival, morphology and GFP signals. (D) Identification of outliers using the Fluidigm SINGuLAR analysis package. Box plot of the expression levels of a set of genes detected in at least half of the samples. Eight samples (red) with median expression (blue or red dot in the box plot) values below the fifteenth percentile (dashed horizontal line) for these genes were removed before further analysis. (E) Principal components analysis (PCA) performed on the 31 cells using the Fluidigm SINGuLAR analysis package. Three main clusters were identified as marked by the ellipses. (F) Hierarchical clustering of the top 500 PCA genes across 31 cells performed in the Fluidigm SINGuLAR package. The three main clusters are labeled in the same color as in E. The green cluster was further divided into two groups with apparent differential gene expression (dark green and light green). Labels I, II, III and IV denote groups. |
| Gene: | |
|---|---|
| Fish: | |
| Anatomical Term: | |
| Stage: | Adult |