Fig. S1
- ID
- ZDB-FIG-160226-27
- Publication
- Wu et al., 2016 - Spatially Resolved Genome-wide Transcriptional Profiling Identifies BMP Signaling as Essential Regulator of Zebrafish Cardiomyocyte Regeneration
- Other Figures
- All Figure Page
- Back to All Figure Page
|
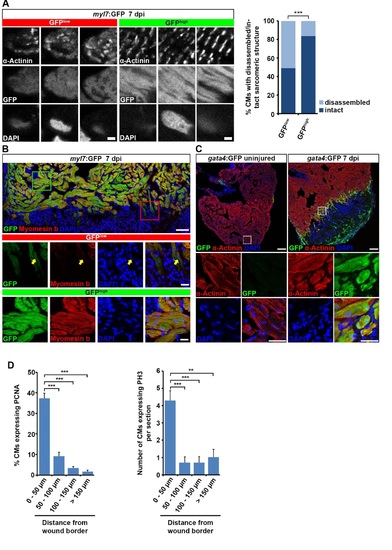
Cardiomyocytes at the wound border dedifferentiate and re-enter the cell cycle. Related to Figure 1. (A) Individual GFPlow and GFPhigh cardiomyocytes were randomly picked on sections of myl7:GFP transgenic hearts and their sarcomeric structure was categorized according to α-Actinin protein arrangement. Striated and disorganized pattern of α-Actinin protein expression was categorized as intact and disassembled sarcomeric structure, respectively. A higher fraction of GFPlow cardiomyocytes display disassembled sarcomeric structures than GFPhigh cardiomyocytes at 7 dpi. Scale bar, 12.5 µm. Fisher’s exact test, p = 0.0002. (B) GFPlow cardiomyocytes show reduced Myomesin b expression (arrow) in myl7:GFP transgenic hearts at 7 dpi. Scale bar, 50 µm (overview) and 25 µm (inset). (C) GFP expression in gata4:GFP transgenic hearts is activated in α-Actinin+ cardiomyocytes at the wound border at 7 dpi. Scale bar, 50 µm (overview) and 12.5 µm (inset). (D) Proliferating cardiomyocytes, as revealed by PCNA and PH3 expression, are mainly localized within 50 µm of the wound border. Average percentage of PCNA and average number of PH3 positive cardiomyocytes at various distances from the wound border is plotted. Error bars represent SEM. Student’s t test, PCNA: p = 0.00002 (0 – 50 µm vs 50 – 100 µm), p = 0.000001 (0 – 50 µm vs 100 – 150 µm), p = 0.0000008 (0 – 50 µm vs >150 µm); PH3: p = 0.0006 (0 – 50 µm vs 50 – 100 µm), p = 0.0006 (0 – 50 µm vs 100 – 150 µm), p = 0.0019 (0 – 50 µm vs >150 µm). |
Reprinted from Developmental Cell, 36(1), Wu, C.C., Kruse, F., Vasudevarao, M.D., Junker, J.P., Zebrowski, D.C., Fischer, K., Noël, E.S., Grün, D., Berezikov, E., Engel, F.B., van Oudenaarden, A., Weidinger, G., Bakkers, J., Spatially Resolved Genome-wide Transcriptional Profiling Identifies BMP Signaling as Essential Regulator of Zebrafish Cardiomyocyte Regeneration, 36-49, Copyright (2016) with permission from Elsevier. Full text @ Dev. Cell