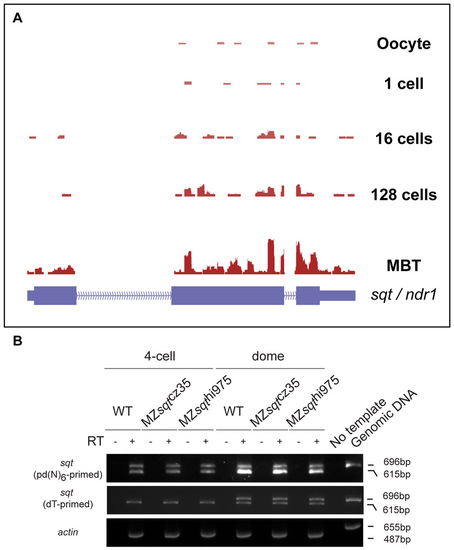
Fig. S1
|
RNA sequencing in early embryos shows non-polyadenylated maternal squint RNA. RNA sequencing of transcripts shows that sqt RNA is not polyadenylated in oocytes and 1-cell embryos, and becomes polyadenylated during early development. (A) RNA sequencing, data normalization, mapping and data analysis are as described previously (Aanes et al., 2011). RNA-seq tags obtained from SOLiD3 sequencing of whole transcriptome libraries from oocytes, 1-cell embryos, 16-cell embryos, 128-cell embryos and 1000-cell (MBT) stage embryos were mapped to Reference Genome assemblies and gene annotation (RefSeq) (UCSC browser Zv9 assembly; danRer7) using Whole Transcriptome Analysis Pipeline version 1.2 (WTAP 1.2) and Bioscope (Applied Biosystems). Tags mapping to all the known RefSeq genes were counted and the mapping was visualized using the browser. Schematic of the sqt (ndr1) locus is shown in blue. (B) RT-PCR using pd(N)6-primed cDNAs (top panel) or oligo(dT)-primed cDNA (middle panel) to detect sqt RNA in 4-cell and dome stage wild-type, MZsqtcz35 and MZsqthi975 embryos. PCR using pd(N)6-primed cDNA shows unspliced (696 bp) and spliced (615 bp) sqt RNA. Spliced sqt product is detected in oligo(dT)-primed cDNA at early stages, and no PCR product is detected in RT and no template controls. actin RT-PCR (bottom panel) and genomic DNA PCR were used as positive controls. |