Fig. 1
- ID
- ZDB-FIG-100223-6
- Publication
- Li et al., 2010 - A systematic approach to identify functional motifs within vertebrate developmental enhancers
- Other Figures
- All Figure Page
- Back to All Figure Page
|
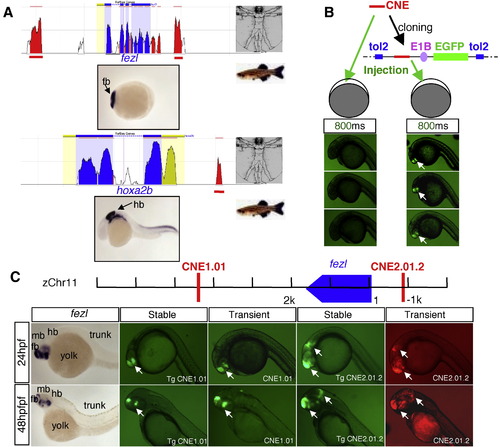
In vivo enhancer detection in zebrafish. (A) A schematic showing identification of CNEs (red peaks) near anterior brain-expressed or posterior brain-expressed genes through comparative bioinformatics analysis using the ECR browser (Ovcharenko et al., 2004) between human and zebrafish. (B) Schematic diagram showing the enhancer detection construct composed of cloning sites for CNEs, E1B basal promoter, and reporter in Tol2 transposon vector, and comparison of two enhancer detection methods. With 800 ms exposure time, the GFP signal is robust in embryos injected with cloned CNE (right), whereas the signal is below detection in embryos injected with PCR product of CNE (left). (C) Comparison of reporter signals in transient versus stable transgenic embryos. CNE1.01 and CNE2.01.2 are identified near the fezl gene. Similar activity is observed between transient (CNE1.01, CNE2.01.2) and stable (Tg CNE1.01, Tg CNE 2.01.2) transgenic embryos. |
Reprinted from Developmental Biology, 337(2), Li, Q., Ritter, D., Yang, N., Dong, Z., Li, H., Chuang, J.H., and Guo, S., A systematic approach to identify functional motifs within vertebrate developmental enhancers, 484-495, Copyright (2010) with permission from Elsevier. Full text @ Dev. Biol.