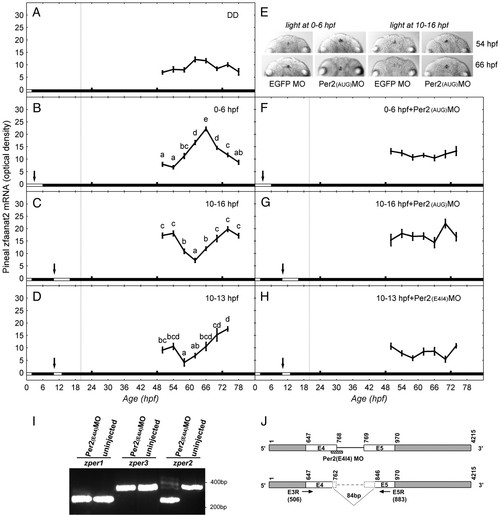
Effect of light and zper2 expression at early embryonic stage on development of pineal zfaanat2 mRNA rhythms. Zebrafish embryos, MO-injected and uninjected, were exposed to light for different periods during the first 16 h of development. During the third and fourth day of development, larvae were collected at 4-h intervals and pineal zfaanat2 mRNA levels were determined by ISH. (A) No rhythm was observed if embryos were placed in darkness at 2 hpf. (B–D) Rhythms of zfaanat2 mRNA appeared in embryos that were exposed to light 0–6 hpf (B), 10–16 hpf (C), or 10–13 hpf (D); note the phase difference induced by the light treatments. (F and G) Microinjection of Per2(AUG)MO at one-cell stage abolished the effect of the 0–6 hpf and 10–16 hpf light treatments (compare B to F and C to G). (H) Microinjection of Per2(E4I4)MO at one-cell stage abolished the effect of the 10–13 hpf light treatments (compare D to H). (E) Representative pineal zfaanat2 signals (dorsal view) at subjective midday (54 hpf) and subjective midnight (66 hpf) of embryos that were exposed to light at 0–6 hpf (Left) or 10–16 hpf (Right) and were injected with Per2(AUG)MO or EGFP MO. Each value represents the average optical density of pineal signal ± SEM in 20–60 embryos. Different letters represent statistically different values (P < 0.05, ANOVA, Tukey’s test). The horizontal bar at the bottom of each panel represents the lighting conditions: white boxes represent the illumination period, and black boxes represent dark. Arrows indicate the beginning of effective illumination. Vertical dashed lines represent the time of pineal gland formation (i.e., earliest detection of photoreceptor cells). (I and J) Per2(E4I4)MO injection altered zper2 mRNA splicing but had no effect on the splicing of zper1 and zper3, as revealed by PCR analysis (I). Sequence analysis of the PCR products indicate that Per2(E4I4)MO caused the skipping of 84 nucleotides in exons 4 and 5 (7 bp in exon 4 and 77 bp in exon 5), which encode a 28-aa fragment in zPER2 PAS A domain (J).
|

