- Title
-
Multi-population GWAS for venous thromboembolism identifies novel loci followed by experimental validation in zebrafish
- Authors
- Wolford, B.N., Zhao, Q.Y., Wu, K.H., Yu, X., Richter, C.E., Bhatta, L., Brumpton, B.M., Desch, K.C., Thibord, F., Klarin, D., Johnson, A.D., Tregouet, D.A., Damrauer, S.M., Smith, N.L., Lo Faro, V., Tsuo, K., Daly, M., Neale, B., Zhou, W., Willer, C.J., Shavit, J.A., Surakka, I.
- Source
- Full text @ Blood Adv
|
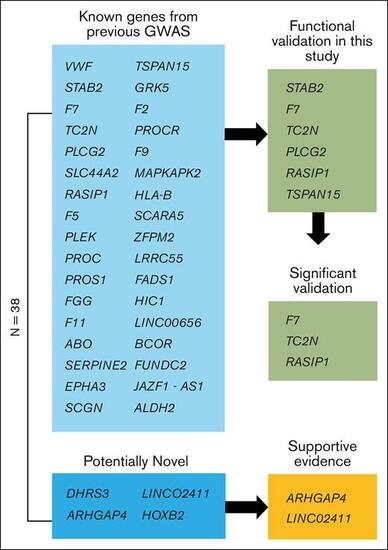
Schematic view of genes. Of the 38 genome-wide significant loci, 4 are potentially novel, and 34 are known from previous GWASs. Of the potentially novel genes, 2 have supportive evidence from recent GWAS meta-analyses.38,39 Six of the previously known genes were functionally validated in this study using a zebrafish model of blood clotting with 3 genes showing supportive evidence (significant validation) as the causal gene for modification of VTE in humans. |
|
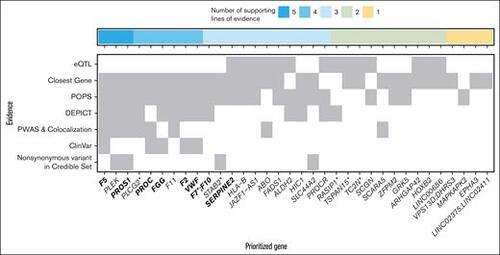
Integrative gene prioritization. Autosomal genome-wide significant loci labeled by prioritized gene (x-axis) with shading for each line of evidence used in the bioinformatics-driven prioritization scheme (y-axis). Genes in bold were on the gold standard list (supplemental Table 7). The 7 lines of evidence evaluated were chosen to cover different mechanisms through which genetic variants contribute to disease risk, for example, regulatory changes vs protein perturbations. For VPS13D;DHRS3, F7;F10, and LINC02375;LINC02411, the genes had equal numbers of supporting lines of evidence. LINC00656 is also known as RP4-737E23.2. rs536995174 had 1 line of evidence each for SERPING1, SLC43A3, SLC43A1, F2, OR5AK4P, and LRRC55 and was excluded from this visualization. Genes with asterisks were selected for follow-up in a functional assay in zebrafish (Figure 3). |
|
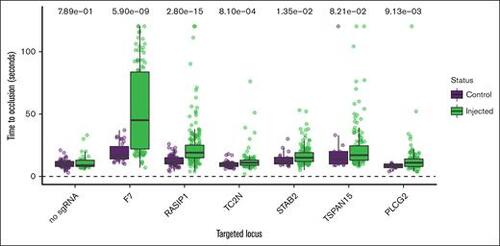
Functional evidence for causal genes in genetically modified zebrafish.P values from Wilcoxon rank sum tests are listed at the top. The y-axis represents the experimental TTO for control and sgRNA-injected zebrafish embryos with the x-axis showing the genes targeted through CRISPR. Injections made without sgRNA served as a negative control. Factor 7 (F7) served as a positive control. |