- Title
-
Colorectal cancer progression to metastasis is associated with dynamic genome-wide biphasic 5-hydroxymethylcytosine accumulation
- Authors
- Murcott, B., Honig, F., Halliwell, D.O., Tian, Y., Robson, J.L., Manasterski, P., Pinnell, J., Dix-Peek, T., Uribe-Lewis, S., Ibrahim, A.E.K., Sero, J., Gurevich, D., Nikolaou, N., Murrell, A.
- Source
- Full text @ BMC Biol.
|
5hmC levels are increased in liver metastasis tissue compared to primary colon tumours in CRC. |
|
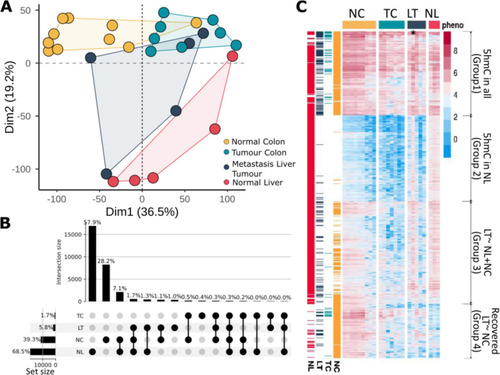
5hmC profiles in liver metastasis and colon are similar. Colour scheme: yellow normal colon tissue from CRC patients (NC); light blue primary tumour in colon (TC); dark blue metastasised to liver tumours (LT); red normal liver (NL). |
|
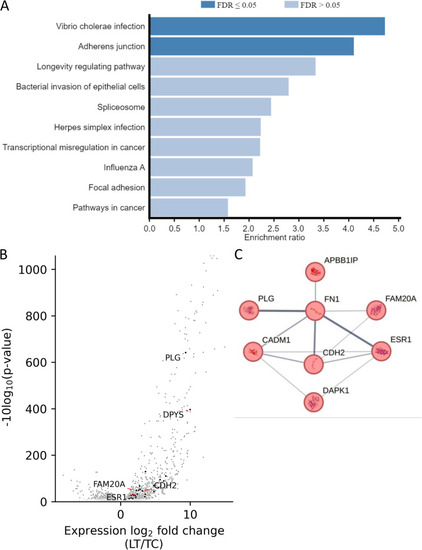
5hmC peaks associated with differential gene expression. |
|
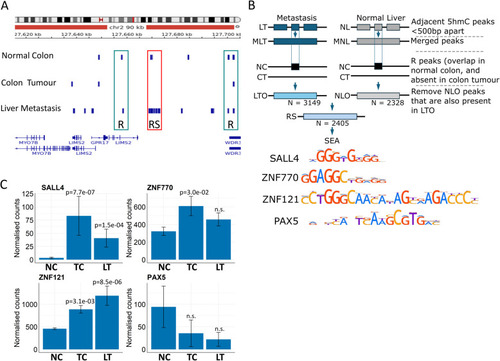
In liver metastasis, 5hmC peaks that were initially present in normal colon are recovered. |