- Title
-
Gastroesophageal circulating tumor cell crosstalk with peripheral immune system guides CTC survival and proliferation
- Authors
- Rossi, T., Valgiusti, M., Puccetti, M., Miserocchi, G., Zanoni, M., Angeli, D., Arienti, C., Pace, I., Bassi, C., Vannini, I., Melloni, M., Bandini, E., Urbini, M., Negrini, M., Bonafè, M., Ferracin, M., Gallerani, G.
- Source
- Full text @ Cell Death Dis.
|
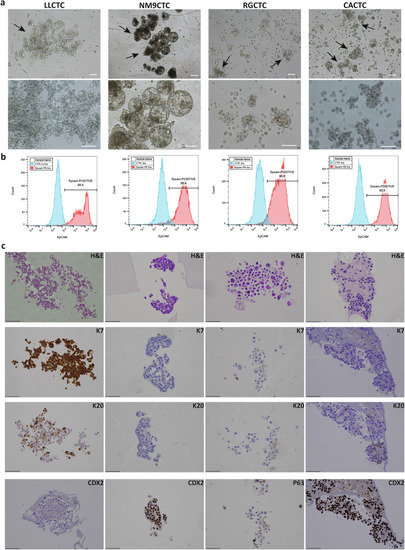
Morphological and phenotypic characteristics of cultured CTCs. |
|
Invasive competence of CTC-derived cell lines in in-vivo zebrafish model. |
|
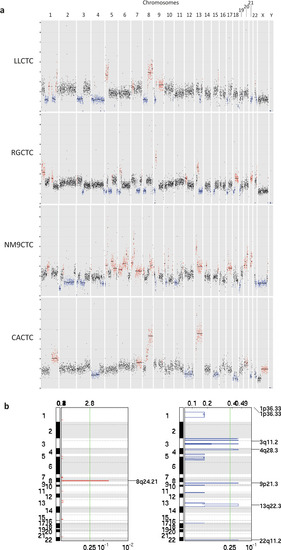
Genomic landscape of the four CTC-derived cell lines. |
|
Gene expression analysis of the four CTC-derived cell lines. |
|
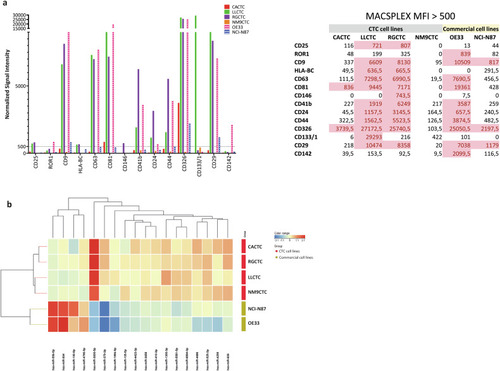
Characterization of extracellular vesicles secreted by cell lines. |
|
Characterization of immune cell differentiation in co-cultures. |