- Title
-
Endothelial tip-cell position, filopodia formation and biomechanics require BMPR2 expression and signaling
- Authors
- Hiepen, C., Benamar, M., Barrasa-Fano, J., Condor, M., Ilhan, M., Münch, J., Hastar, N., Kerkhoff, Y., Harms, G.S., Mielke, T., Koenig, B., Block, S., Rocks, O., Abdelilah-Seyfried, S., Van Oosterwyck, H., Knaus, P.
- Source
- Full text @ Commun Biol
|
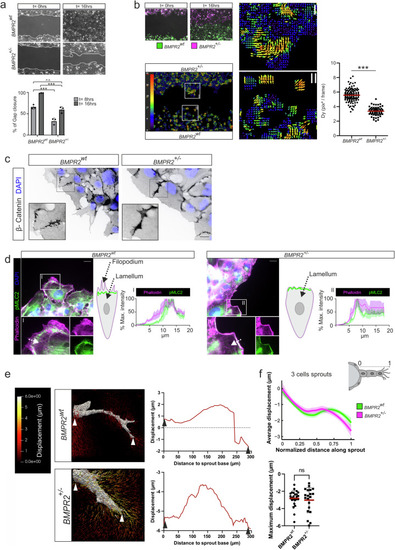
BMPR2 is required for CVP sprouting angiogenesis and EC filopodia formation, contributing to the endothelial tip cell phenotype. |
|
BMPR2 is required for polarized EC migration, regulates spatial actomyosin organization at the EC leading edge and organizes 3D pulling force distribution at the sprout front during angiogenic sprouting in fibrin ECM. |
|
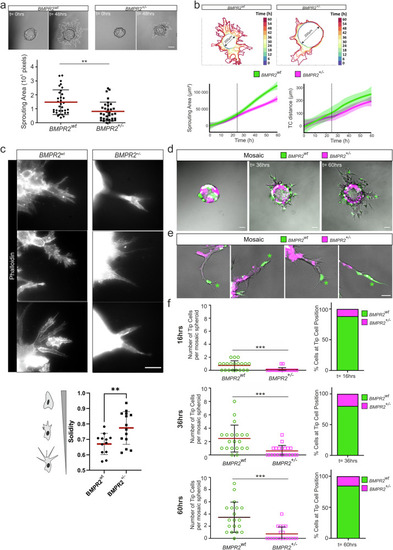
BMPR2 is required for efficient sprouting and tip cell (TC) position in 3D spheroid assays. |
|
BMPR2 promotes CDC42 activity at the plasma membrane of ECs via the PI3K-CDC42 signaling axis. |
|
Graphical summary. Endothelial cells form filopodia to sense their environments and facilitate chemotaxis-induced migration. In 2D migration assays, BMPR2-deficient cells fail to form filopodia and display an exacerbated collective migration behavior upon lack of BMPR2 expression which impedes with their efficient forward movement. We also found an increased co-localization of actomyosin together with cortical actin at the leading edge of BMPR2 deficient cells. Also, focal adhesion formation is altered. In 3D sprouting assays, BMPR2 deficiency stalls sprouting and abrogates ECs from acquiring the tip cell (TC) position. Wildtype and BMPR2 deficient sprouts perform pulling which is dominated by the tip-cell (TC), while we find no general change in overall pulling forces between BMPR2wt and BMPR2+/- sprouts. In spheroid sprouting assays as mosaic, loss of BMPR2 expression by the TC identifies |