- Title
-
MRAP2a Binds and Modulates Activity and Localisation of Prokineticin Receptor 1 in Zebrafish
- Authors
- Fullone, M.R., Maftei, D., Vincenzi, M., Lattanzi, R., Miele, R.
- Source
- Full text @ Int. J. Mol. Sci.
|
|
|
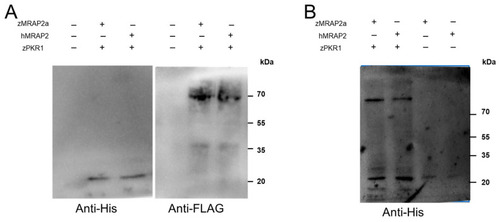
Interaction of zMRAP2a and hMRAP2 isoforms with zPKR1. ( |
|
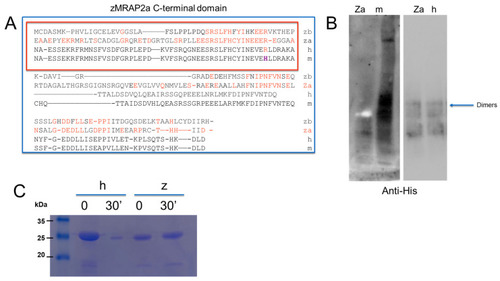
Biochemical analysis of the zCT-MRAP2a domain. ( |
|
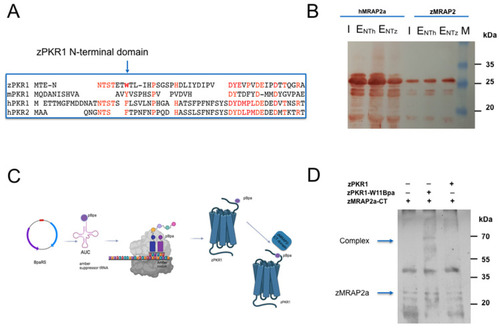
Role of N-Terminal region of zPKR1 for zMRAP2a and hMRAP2 interaction. ( |
|
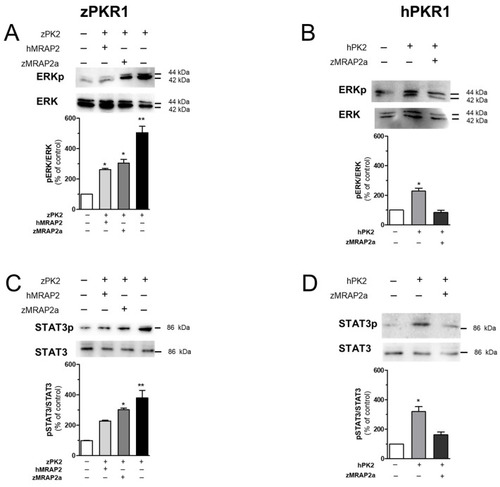
Analysis of ERK and STAT3 activation in CHO cells. ( |
|
( |