- Title
-
Zebrafish Avatars: Toward Functional Precision Medicine in Low-Grade Serous Ovarian Cancer
- Authors
- Fieuws, C., Bek, J.W., Parton, B., De Neef, E., De Wever, O., Hoorne, M., Estrada, M.F., Van Dorpe, J., Denys, H., Van de Vijver, K., Claes, K.B.M.
- Source
- Full text @ Cancers
|
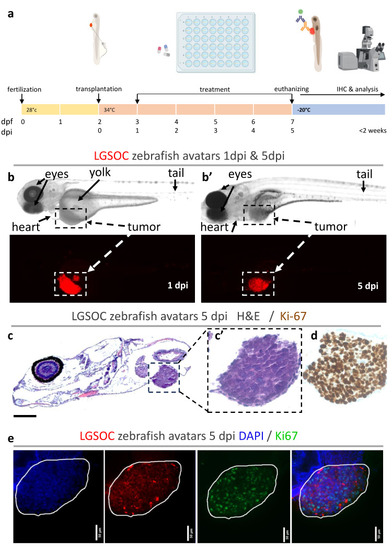
Establishment and analysis of zebrafish xenografts from LGSOC cells. ( |
|
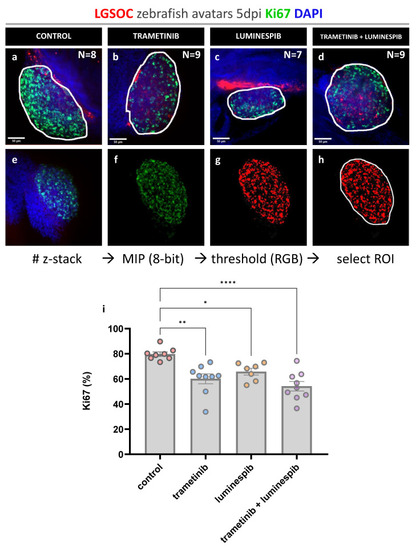
Whole mount immune fluorescence staining for Ki67. Vybrant CM-DiI labeled LGSOC cells (red), nuclei stained with DAPI (blue) and anti Ki-67 (green). ( |
|
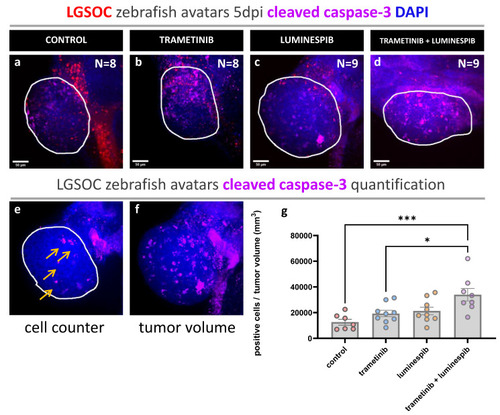
Whole mount immune fluorescence staining cleaved caspase-3. Vybrant CM-DiI labeled LGSOC cells (red), nuclei stained with DAPI (blue) and anti-cleaved caspase-3 (violet). ( |