- Title
-
Klebsiella pneumoniae alters zebrafish circadian rhythm via inflammatory pathways and is dependent on light cues
- Authors
- Ding, H., Chen, X.C., Wan, L., Zhang, Y.Y., Rui, X.H., He, T., Liu, J., Shang, Z.B.
- Source
- Full text @ Heliyon
|
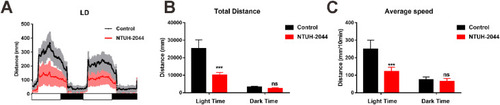
Locomotor ability of zebrafish exposed to Klebsiella pneumoniae decreased under 14 h/10 h light-dark (L/D) conditions. (A) Locomotor ability of juvenile fish in the control and NTUH-2044 groups under L/D conditions. (B) Total travel distance of juvenile zebrafish in light and dark time periods in the control and NTUH-2044 groups. (C) Average swimming speed of juvenile zebrafish in light and dark time periods in the control and NTUH-2044 groups. Each experiment was repeated in triplicate. Data are expressed as the mean ± standard deviation (SD). Data were analyzed using Student's t-test. (***P < 0.001, ns: not significant). |
|
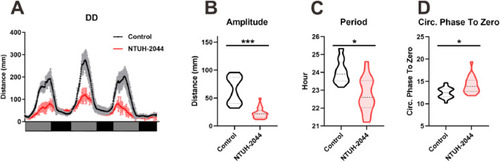
Changes in behavior of Klebsiella pneumoniae-infected zebrafish under continuous darkness (D/D) conditions. (A) Behavioral rhythm analysis of juvenile fish in the control and NTUH-2044 groups under D/D conditions. (B) Amplitude comparison of juvenile zebrafish in the control and NTUH-2044 groups under D/D conditions. (C) Period comparison of juvenile zebrafish in the control and NTUH-2044 groups under D/D conditions. (D) Phase comparison of juvenile zebrafish in the control and NTUH-2044 groups under D/D conditions. Each experiment was repeated three times. Data are expressed as the mean ± standard deviation (SD). Data were analyzed using Student's t-test. (*P < 0.05, **P < 0.01, ***P < 0.001). |
|
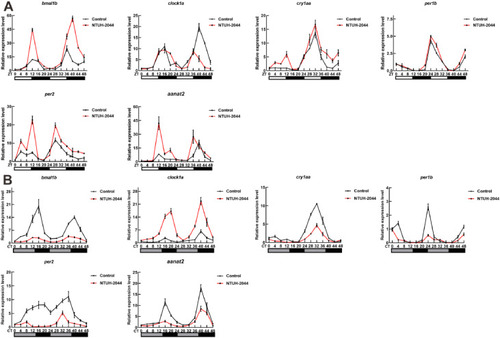
Expression of core circadian rhythm genes in Klebsiella pneumoniae-infected zebrafish. Expression levels of (A) bmal1b, clock1a, cry1aa, per1b, per2, and aanat2 in the control and NTUH-2044 groups under light/dark (L/D) conditions. Expression levels of (B) bmal1b, clock1a, cry1aa, per2, per1b, and aanat2 in the control and NTUH-2044 treatment groups under D/D conditions. Each experiment was repeated three times. Data are expressed as the mean ± standard deviation (SD). Data were analyzed using Student's t-test. (*P < 0.05, **P < 0.01, ***P < 0.001). |
|
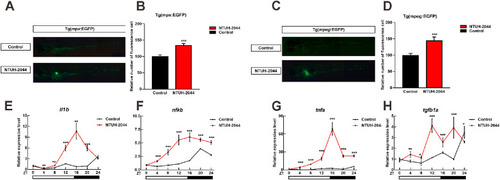
Inflammatory cell and gene expression levels in zebrafish infected with Klebsiella pneumoniae and treated under light/dark (L/D) conditions. (A) Neutrophil recruitment of Tg(mpx:EGFP) transgenic zebrafish in the control and NTUH-2044 treatment groups under L/D conditions. (B) Number of neutrophils in Tg(mpx:EGFP) transgenic zebrafish compared with those in the control and NTUH-2044 groups under L/D conditions. (C) Macrophage recruitment of Tg(mpeg:EGFP) transgenic zebrafish in the control and NTUH-2044 groups under L/D conditions. (D) Number of macrophages in Tg(mpeg:EGFP) transgenic zebrafish compared with those in the control and NTUH-2044 groups under L/D conditions. qRT-PCR analysis of inflammation-related genes il1b (E), nfkb (F), tnfa (G), and tgfb1a (H) in the control and NTUH-2044 groups under L/D conditions. Each experiment was repeated three times. Data are expressed as the mean ± standard deviation (SD). Data were analyzed using Student's t-test. (ANOVA) (*P < 0.05, **P < 0.01, ***P < 0.001). |
|
Anti-inflammatory drugs reduce Klebsiella pneumoniae-induced inflammation. Zebrafish were infected with NTUH-K2044 for 48 hpf, followed by treatment with the anti-inflammatory drug G6PDi-1, and inflammation was evaluated at 5 days post-fertilization (dpf). (A) Neutrophil recruitment in Tg(mpx:EGFP) transgenic zebrafish in the control, NTUH-2044, and NTUH-2044+G6PDi-1 treatment groups. (B) Number of neutrophils in Tg(mpx:EGFP) transgenic zebrafish in the control, NTUH-2044, and NTUH-2044+G6PDi-1 treatment groups. (C) Macrophage recruitment of Tg(mpx:EGFP) transgenic zebrafish in the control, NTUH-2044, and NTUH-2044+G6PDi-1 treatment groups. (D) Number of macrophages in Tg(mpx:EGFP) transgenic zebrafish in the control, NTUH-2044, and NTUH-2044+G6PDi-1 treatment groups. qRT-PCR analysis of inflammation-related genes il1b (E), nfkb (F), tnfα (G), and tgfb1a (H) in the control, NTUH-2044, and NTUH-2044+G6PDi-1 treatment groups. Each experiment was performed in triplicate. Data are expressed as the mean ± standard deviation (SD). Data were analyzed by Student's t-test.(ANOVA) (**P < 0.01,***P < 0.001). |
|
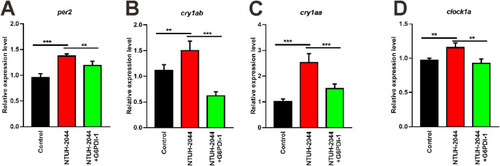
Anti-inflammatory drug rescue of Klebsiella pneumoniae-disrupted circadian rhythm genes. Zebrafish were infected with NTUH-K2044 for 48 hpf, followed by treatment with the anti-inflammatory drug G6PDi-1, and the expression of genes associated with circadian rhythm were evaluated in different groups of zebrafish at 5 dpf. qRT-PCR analysis of zebrafish clock genes per2 (A), cry1ab (B), cry1aa (C), and clock1a (D) in the control, NTUH-2044, and NTUH-2044+G6PDi-1 treatment groups. Each experiment was performed in triplicate. Data are expressed as the mean ± standard deviation (SD). Data were analyzed using Student's t-test. (**P < 0.01, ***P < 0.001). |
|
Proposed scheme of how Klebsiella pneumoniae alters zebrafish circadian rhythm via inflammatory pathways dependent on light cues. |