- Title
-
SUDAZFLNC - a curated and searchable online database for zebrafish lncRNAs, mRNAs, miRNAs, and circadian expression profiles
- Authors
- Mishra, S.K., Wang, H.
- Source
- Full text @ Comput Struct Biotechnol J
|
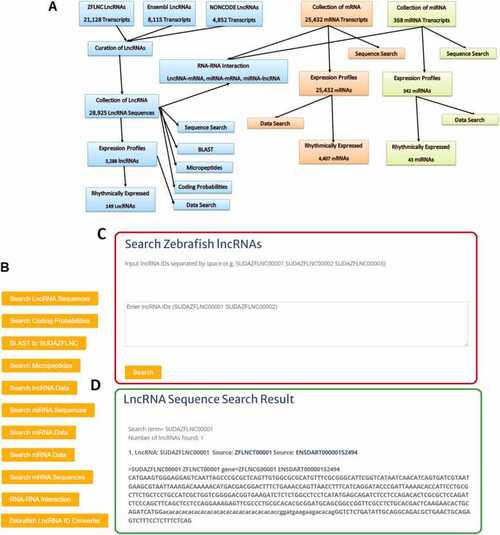
Database development flowchart, features, and searching zebrafish lncRNA sequences. (A) Flow chart of the development of the SUDAZFLNC database in the study. (B) List of all features supported by the database. (C) Input of identifiers to search the lncRNA sequence. (D) Output of lncRNA search query. |
|
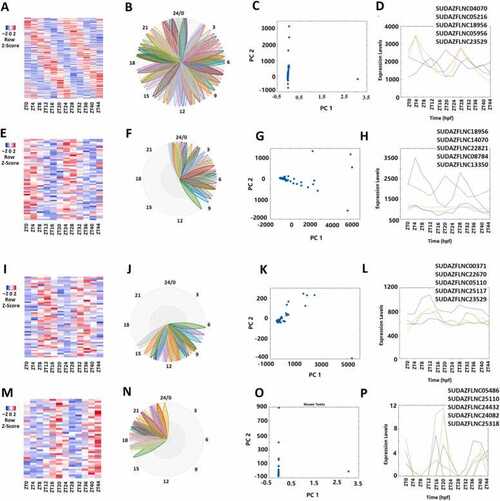
Analyses of 149 rhythmically expressed zebrafish lncRNAs. (A-D) Analysis of all the 149 rhythmically expressed zebrafish lncRNAs: Heat map (A) of all the 149 rhythmically expressed lncRNAs, BioDare2 phase plots of all lncRNAs (B), PCA analyses of all lncRNAs with variances of PC1 99.96% and PC2 0.02% (C), and expression profiles of the representative lncRNAs (D). (E-H) Analysis of 56 morning lncRNAs: Heat map of the 56 morning lncRNAs (E), BioDare2 phase plots of morning lncRNAs (F), PCA analyses of the morning lncRNAs with variances of PC1 93.94% and PC2 3.97% (G), and expression profiles of the representative lncRNAs (H). (I-L) Heat map of the 52 evening lncRNAs (I), BioDare2 phase plots of evening lncRNAs (J), PCA analyses of the evening lncRNAs with variances of PC1 98.53% and PC2 0.79% (K), and expression profiles of the representative lncRNAs (L). (M-P) Heat map of the 41 night lncRNAs (M), BioDare2 phase plots of the night lncRNAs (N), PCA analyses of the night lncRNAs with variances of PC1 99.99% and PC2 0.01% (O), and expression profiles of the representative lncRNAs (P). |
|
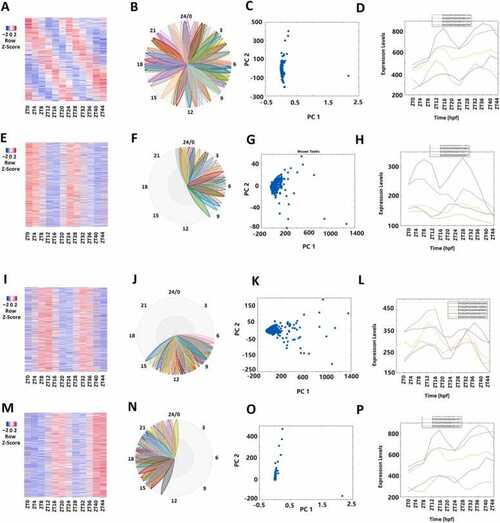
Analyses of 4047 rhythmically expressed zebrafish mRNAs. (A-D) Analysis of all the 4047 rhythmically expressed zebrafish mRNAs: Heat map (A) of all the 4047 rhythmically expressed mRNAs, BioDare2 phase plots of all mRNAs (B), PCA analyses of all mRNAs with variances of PC1 99.56% and PC2 0.19% (C), and expression profiles of the representative mRNAs (D). (E-H) Analysis of 1817 morning mRNAs: Heat map of the 1817 morning mRNAs (E), BioDare2 phase plots of morning mRNAs (F), PCA analyses of the morning mRNAs with variances of PC1 97.82% and PC2 1.04% (G), and expression profiles of the representative mRNAs (H). (I-L) Heat map of the 1128 evening mRNAs (I), BioDare2 phase plots of evening mRNAs (J), PCA analyses of the evening mRNAs with variances of PC1 97.18% and PC2 1.75% (K), and expression profiles of the representative mRNAs (L). (M-P) Heat map of the 1426 night mRNAs (M), BioDare2 phase plots of the night mRNAs (N), PCA analyses of the night mRNAs with variances of PC1 99.82% and PC2 0.13% (O), and expression profiles of the representative mRNAs (P). |
|
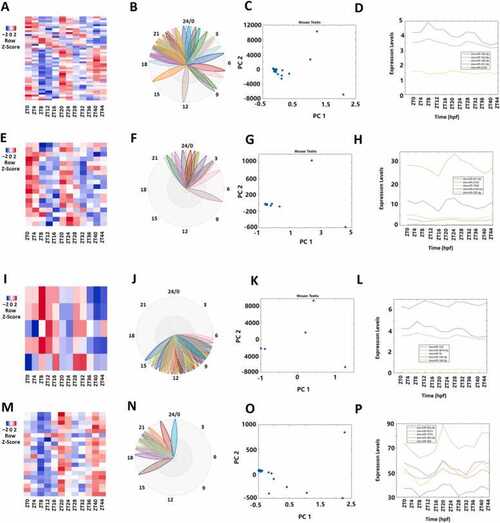
Analyses of 43 rhythmically expressed zebrafish miRNAs. (A-D) Analysis of all the 4047 rhythmically expressed zebrafish miRNAs: Heat map (A) of all the 43 rhythmically expressed miRNAs, BioDare2 phase plots of all miRNAs (B), PCA analyses of all miRNAs with variances of PC1 99.73% and PC2 0.21% (C), and expression profiles of the representative miRNAs (D). (E-H) Analysis of 18 morning miRNAs: Heat map of the 18 morning miRNAs (E), BioDare2 phase plots of morning miRNAs (F), PCA analyses of the morning miRNAs with variances of PC1 99.94% and PC2 0.04% (G), and expression profiles of the representative miRNAs (H). (I-L) Heat map of the 5 evening miRNAs (I), BioDare2 phase plots of evening miRNAs (J), PCA analyses of the evening miRNAs with variances of PC1 99.56% and PC2 0.41% (K), and expression profiles of the representative miRNAs (L). (M-P) Heat map of the 20 night miRNAs (M), BioDare2 phase plots of the night miRNAs (N), PCA analyses of the night miRNAs with variances of PC1 99.85% and PC2 0.10% (O), and expression profiles of the representative mRNAs (P). |
|
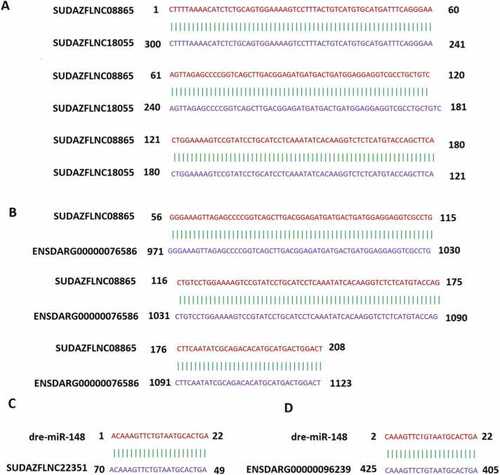
RNA-RNA interaction and target prediction. LncRNA-lncRNA target prediction: binding of lncRNA SUDAZFLNC08865 to lncRNA SUDAZFLNC18055 (A), lncRNA-mRNA target prediction: binding of lncRNA SUDAZFLNC08865 to mRNA ENSDARG00000076586 (B), miRNA-lncRNA target prediction: binding of miRNA dre-miR-148 to lncRNA SUDAZFLNC22351 (C), and miRNA-mRNA target prediction: binding of miRNA dre-miR-148 to mRNA ENSDARG00000096239 (D). |