- Title
-
Experimental evolution of Staphylococcus aureus in macrophages: dissection of a conditional adaptive trait promoting intracellular survival
- Authors
- Alves, J., Vrieling, M., Ring, N., Yebra, G., Pickering, A., Prajsnar, T.K., Renshaw, S.A., Fitzgerald, J.R.
- Source
- Full text @ MBio
|
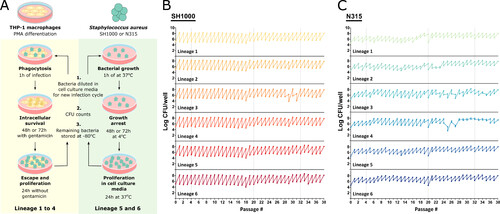
An experimental model of S. aureus evolution in human macrophages. (A) Schematic representation of the long-term S. aureus THP-1 macrophage infection protocol. For each infection cycle, THP-1 macrophages were differentiated by PMA stimulation (as described in Materials and Methods) and infected with Lineages 1–4 of N315 or SH1000 for 1 h. Bacteria were kept under intracellular selective pressure by incubating the culture with a low dose of gentamicin for 48 h or 72 h. Antibiotics were then removed and the bacteria were allowed to escape and multiply in the extracellular space for 24 h. As a control, N315 and SH1000 Lineages 5 and 6 were grown in macrophage culture media alone. The bacteria were incubated at 37°C for 1 h and then incubated at 4°C for 48 h to 72h to mimic the growth restriction of the antibiotic step of Lineages 1–4. The culture was then moved to the 37°C incubator for 24 h. At the end of the cycle, the cell culture media was collected and used for a new infection cycle, bacterial quantification, and colony analysis in TSA and blood agar plates, and the remainder was stored at −80°C in glycerol. Figure made using Servier Medical Images. (B and C) Amount of bacteria used for infection at the beginning of each infection cycle and recovered at the end of each cycle, after 24 h at 37°C without antibiotics. Lineages 1–4 evolved in the presence of THP-1 macrophages and Lineages 5–6 in macrophage culture media alone. Total of 38 cycles. |
|
Mutations acquired during S. aureus sequential infections of macrophages. (A) Distribution of different types of mutations across the genomes and genes affected, colored by type of mutation. Mutations in isolates from passage cycles 4, 20, and 32 are plotted in the inner, middle, and outer circles, respectively. (B) Frequency of the types of mutations (from the 90 Illumina sequenced isolates per strain). (C) List of mutations found in more than three isolates in the passage cycle 4 (P4), 20 (P20), and 32 (P32), colored by the number of isolates with the mutation (from the five sequences per lineage, passage cycle, and strain). Synonymous mutations in italic. |
|
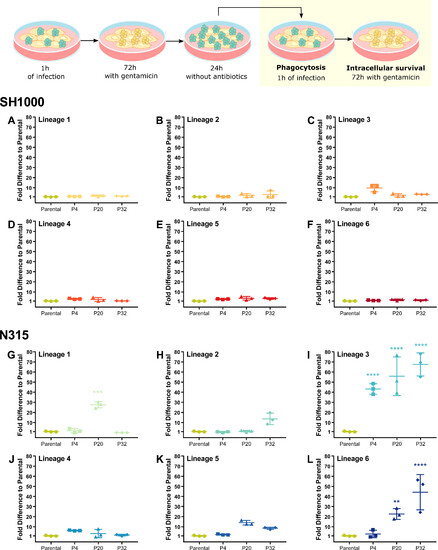
N315 Lineage 3 isolates exhibit increased fitness to survive THP-1 macrophage killing. THP-1 macrophages were infected with SH1000 (A–F) and N315 (G–L) parental strains and isolates from passage cycles 4 (P4), 20 (P20), and 32 (P20) from Lineages 1 to 6 after one cycle of THP-1 infection. THP-1 macrophages were infected with S. aureus at MOI 1 for 1 h, gentamicin was added for 72 h, and bacteria were allowed to escape and divide in the macrophage cell culture media prior to a new cycle of infection. New cycle: after 1 h of infection, the media was replaced with media with gentamicin. Bacterial survival was evaluated by comparing bacterial levels after 72 h inside the macrophages to bacterial levels after 1 h of phagocytosis. One-way ANOVA with Holm-Sidak’s multiple comparisons test to Parental. **P < 0.01, ***P < 0.001, ****P < 0.0001. |
|
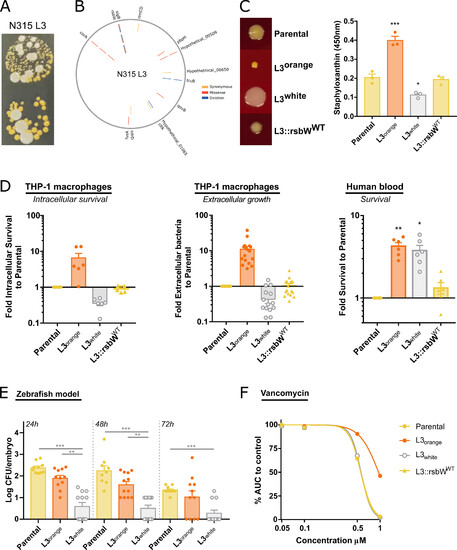
S. aureus N315 Lineage 3 is a hyper-pigmented SCV variant with an unstable colony phenotype. (A) Images of the unstable colony phenotype of N315 L3 passage cycle 32, with hyper-pigmented SCV and large white colonies. (B) Mutations identified in N315 L3 isolates from passage cycle 4 (inner circle), 20 (middle circle), and 32 (outer circle). Mutations in corA and rsbW were identified and maintained since passage cycle 4. (C) Reversion of rsbW mutation to WT in N315 L3 isolate reverts the colony phenotype. Photos of colony morphology from N315 parental, L3 orange, L3 white, and L3 with the rsbW mutation reverted to WT in THA incubated for 48 h at 37°C. Staphyloxanthin quantification after methanol extraction. (D) Bacteria grown overnight at 37°C in nutrient-rich media prior to infection. L3 orange and L3 white isolates showed different survival in human blood, intracellular survival (after 72 h with gentamicin), and extracellular growth in THP-1 cultures (after 24 h without antibiotics) to parental strain and L3::rsbWWT. (E) L3 white achieves lower bacterial levels than parental or L3 orange in a zebrafish embryo infection model. (F) L3 orange isolate has an intermediate vancomycin resistance phenotype. Bacteria were grown in TSB in the presence of increasing concentrations of vancomycin and the AUC of growth curves calculated. The AUC of each bacterial strain at the different vancomycin concentrations was normalized to growth curves in the absence of the antibiotic. Each point represents a biological replication. One-way ANOVA with Tukey’s multiple comparisons. *P < 0.05, **P < 0.01, ***P < 0.001. |
|
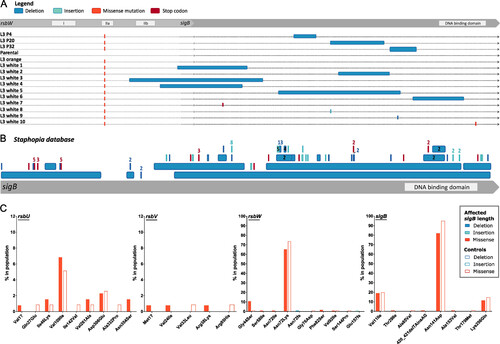
Loss of SCV phenotype in N315 L3 isolates associated with SigB inactivation. (A) Inactivation of sigB was detected in isolates from passage cycles 4, 20, and 32, and in single L3 white isolates. (B) Identification of sigB truncations and deletions among publicly available S. aureus sequences. Representation of deletions (dark blue), insertions (light blue), and early-stop codons (red) in the 134 S. aureus genomes from the Staphopia database with changes in sigB length. The mutation is numbered when identified more than once. (C) Identification of mutations in the sigB operon among clinical S. aureus isolates exclusively associated with SigB-inactivation. Frequency of specific mutations in sigB operon genes among S. aureus clinical isolates with a SigB-inactivation (as in 5B) compared with 118 controls. |