- Title
-
An Efficient Vector-Based CRISPR/Cas9 System in Zebrafish Cell Line
- Authors
- Ye, X., Lin, J., Chen, Q., Lv, J., Liu, C., Wang, Y., Wang, S., Wen, X., Lin, F.
- Source
- Full text @ Mar. Biotechnol.
|
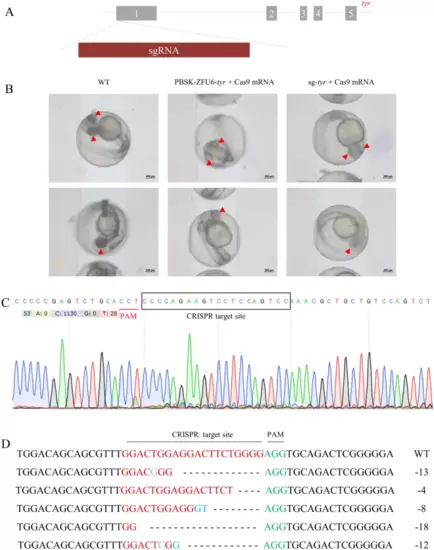
The effectiveness of ZFU6 promoter for gene editing in zebrafish embryos. A Genomic DNA structure and sgRNA site of tyr. B Diagram showing the phenotype caused by mutation of the tyr gene (arrows), scale bar = 200 Ám. C Sequencing of the tyr gene from embryos injected with PBSK-ZFU6-tyr plasmid and Cas9 mRNA. D The mutation type of tyr gene. Dash represents the deleted base, red letters indicate the CRISPR target site, green letters indicate the PAM sites and blue letters indicate the inserted base |
|
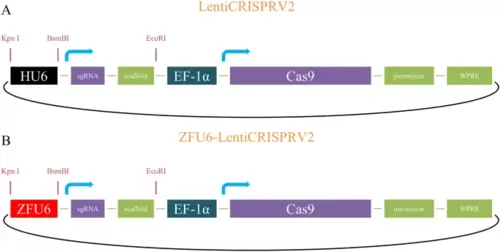
Simplified diagram of the ZFU6-LentiCRISPRV2 vector adapted from mammal LentiCRISPRV2 vector for gene editing in zebrafish cell. A Diagram of mammal LentiCRISPRV2 vector. B Diagram of ZFU6-LentiCRISPRV2 vector. Colors are represented the names of the mainly regulatory elements including RNA polymerase III promoter (HU6/ZFU6), short EF1alpha (EF1?) promoter, single guide RNA (sgRNA), sgRNA scaffold (scaffold), Cas9 protein (Cas9), and the antibiotic resistance cassette (puromycin) |
|
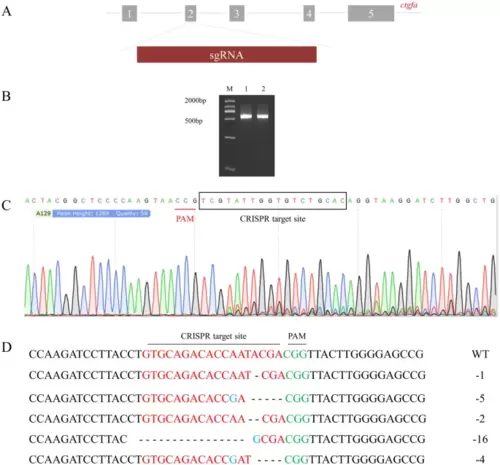
ZFU6-LentiCRISPRV2-ctgfa mediated gene editing in zebrafish PAC2 cell. A Genomic DNA structure and sgRNA site of ctgfa. B PCR products of ctgfa. M: DL2000 DNA marker; 1: cells transfected with ZFU6-LentiCRISPRV2; 2: cells transfected with ZFU6-LentiCRISPRV2-ctgfa. C Sequencing of ctgfa gene from the puromycin-selection cells transfected with ZFU6-LentiCRISPRV2-ctgfa. D The mutation type of ctgfa gene. Dash represents the deleted base, red letters indicate the CRISPR target site, green letters indicate the PAM sites and blue letters indicate the inserted base |
|
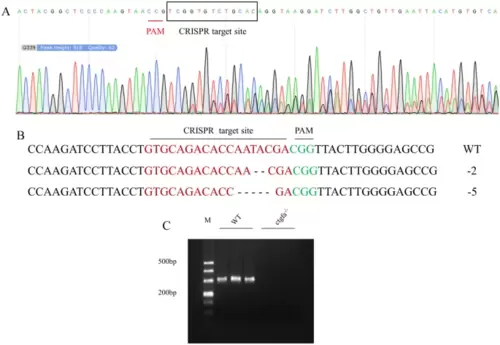
Characterization of a ctgfa knock-out zebarfish PAC2 cell line. A Sequencing of ctgfa gene from the single clone screening cells transfected with ZFU6-LentiCRISPRV2-ctgfa. B The mutation type of ctgfa gene from the single clone screening cells. Dash represents the deleted base, red letters indicate the CRISPR target site, green letters indicate the PAM sites. C RT-PCR products of ctgfa. M: DL500 DNA marker |
|
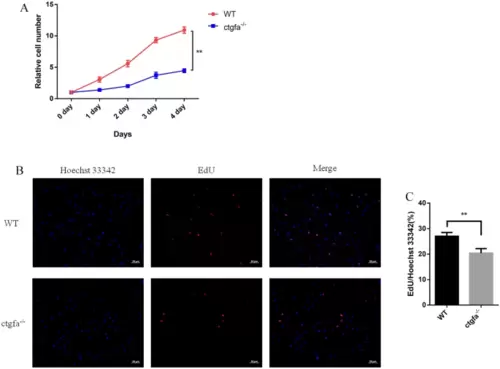
Effect of ctgfa knock out on the growth and proliferation of PAC2 cell. A knockout of ctgfa dramatically reduced PAC2 cell growth, as shown by CCK-8 assay. B, C Knockout of ctgfa led to a decrease in the number of PAC2 cell in S phase as shown by EdU assay. Error bars present means ▒ SD (n = 3); **p < 0.01 |
|
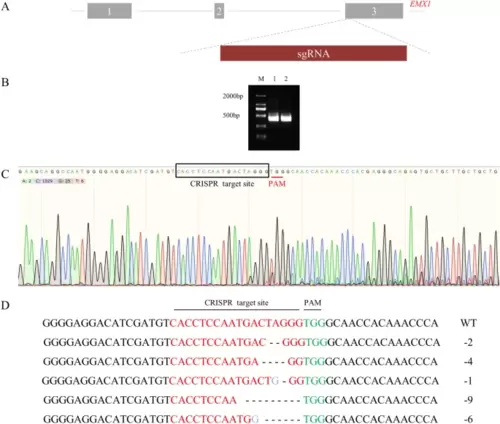
ZFU6-LentiCRISPRV2-EMX1 mediated gene editing in 293 T cell. A Genomic DNA structure and sgRNA site of EMX1. B PCR products of EMX1. M, DL2000 DNA marker; 1, cells transfected with ZFU6-LentiCRISPRV2; 2, cells transfected with ZFU6-LentiCRISPRV2-EMX1. C Sequencing of the EMX1 gene from cells with ZFU6-LentiCRISPRV2-EMX1 transfection and puromycin selection for 7 days. D The mutation type of EMX1 gene. Dash represents the deleted base, red letters indicate the CRISPR target site, green letters indicate the PAM sites and blue letters indicate the inserted base |