- Title
-
Establishment of a zebrafish inbred strain, M-AB, capable of regular breeding and genetic manipulation
- Authors
- Sadamitsu, K., Velilla, F., Shinya, M., Kashima, M., Imai, Y., Kawasaki, T., Watai, K., Hosaka, M., Hirata, H., Sakai, N.
- Source
- Full text @ Sci. Rep.
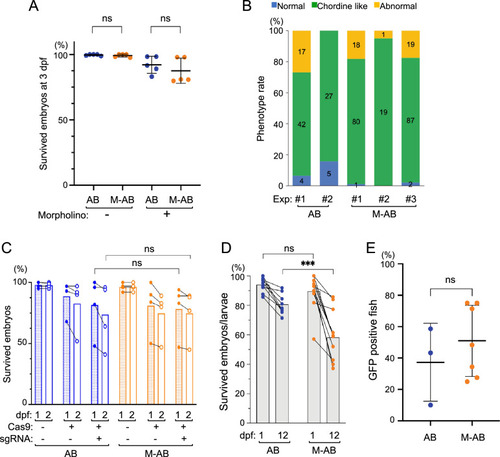
|
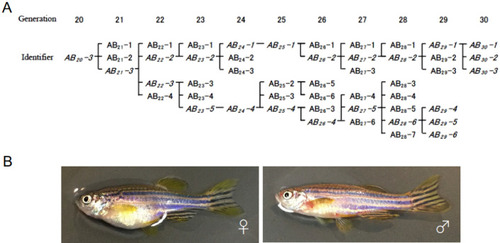
Pedigree of the M-AB strain. ( |
|
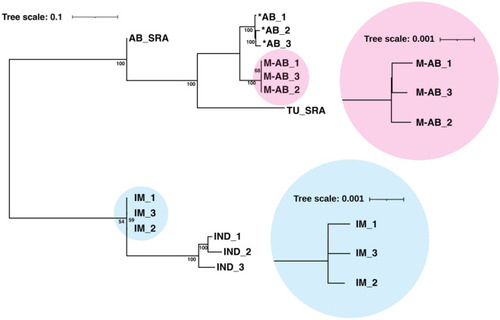
Phylogenetic tree of the M-AB and IM strains. Phylogenetic relationships of the M-AB, *AB, IM and IND strains with open data for AB_SRA and TU_SRA. This tree used 10,000 bootstrap replicates. Coefficients represent bootstrap values for tree nodes. Note that inbred strains have shorter branches than the other strains. The length of the branch indicates the diversity of that strain. |
|
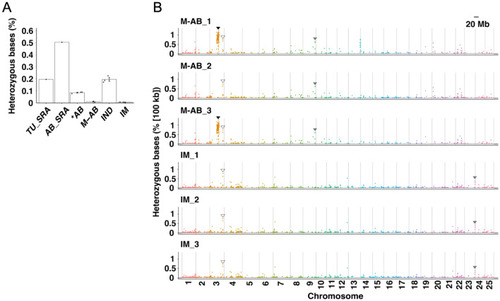
Genomic characteristics of the M-AB and IM strains. ( |
|
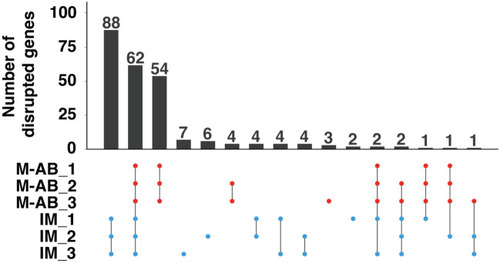
Number of genes potentially disrupted in the M-AB and IM strains. The homozygous SNPs that create nonsense codons or disrupt initiation codons, termination codons or 2-bp splicing junctions within protein coding genes were evaluated as gene disrupting SNPs. The upset plots indicate the overlap of disrupted genes in three M-AB and three IM fish. |
|
Survival of M-AB embryos was hardly affected by microinjections. ( |