- Title
-
Optimal tagging strategies for illuminating expression profiles of genes with different abundance in zebrafish
- Authors
- Liu, J., Li, W., Jin, X., Lin, F., Han, J., Zhang, Y.
- Source
- Full text @ Commun Biol
|
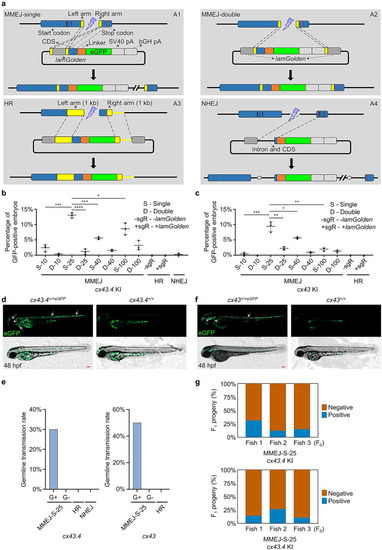
S-25 donor ensures efficient MMEJ-mediated KI in zebrafish. |
|
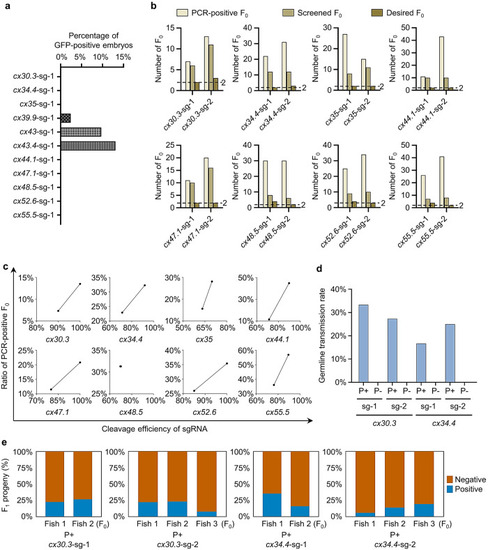
Evaluation of the S-25 strategy by tagging all zebrafish |
|
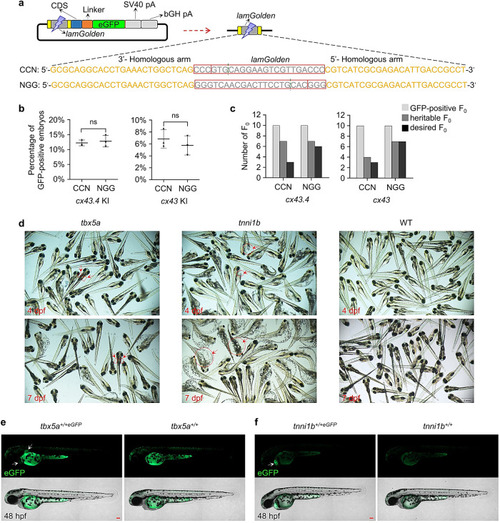
Reducing non-homologous residues introduced by |
|
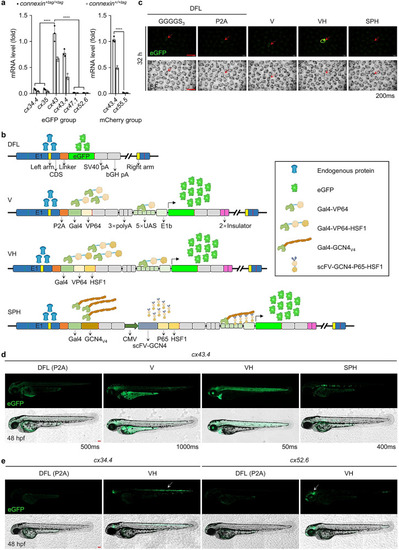
Fluorescent signals can be dramatically amplified by the VH strategy. |
|
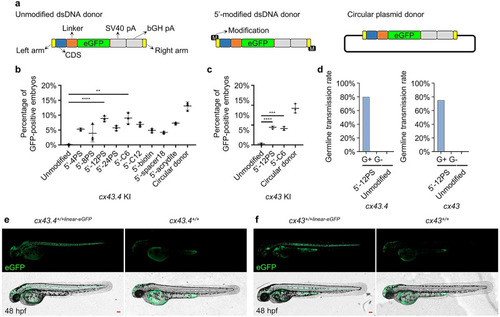
Establishment of 5’-end-modified dsDNA mediated KI system based on S-NGG-25 KI strategy. |