|
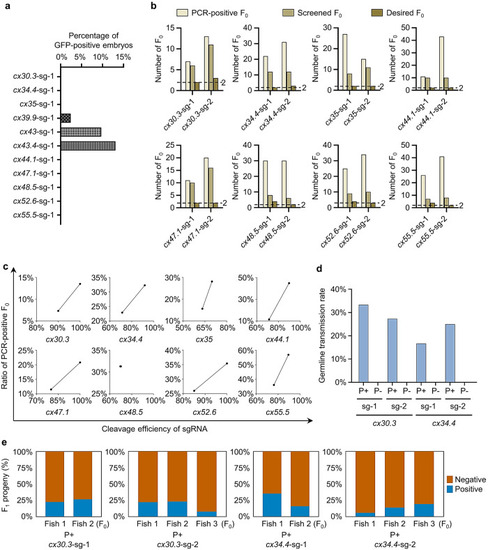
Evaluation of the S-25 strategy by tagging all zebrafish connexins. a Percentages of GFP-positive F0 embryos after tagging cx30.3, cx34.4, cx35, cx39.9, cx43, cx43.4, cx44.1, cx47.1, cx48.5, cx52.6, or cx55.5. S-25 strategy was performed and GFP-positive F0 embryos were counted at 48 hpf. At least 200 embryos were analyzed for each gene. b Number of the desired F0 for cx30.3, cx34.4, cx35, cx44.1, cx47.1, cx48.5, cx52.6, or cx55.5 KI. Genomic DNAs were extracted from the caudal fins of 96 injected F0 zebrafish (except for cx55.5, the genomic DNAs of which were from 72 F0 zebrafish) and used for 5’-junction PCR. PCR-positive F0 are those positive for 5’-junction PCR. Screened F0 are the PCR-positive F0 outbred with WT zebrafish. Desired F0 are the screened F0 whose F1 offspring had precisely repaired 5’-junction. Two high-efficiency sgRNAs were chosen for each gene KI (sg-1 and sg-2). c Correlation between the cleavage efficiencies of sgRNAs and the ratios of PCR-positive F0. The cleavage efficiency of a sgRNA was evaluated by PCR product enzymatic digestions at 24-h post injection or software analysis by the TIDE website. d Germline transmission rates of PCR-positive F0 and PCR-negative F0 for cx30.3 and cx34.4 KI. “P+”, PCR-positive F0; “P-”, PCR-negative F0. Two high-efficiency sgRNAs were used for KI at either locus. At least 6 F0 were analyzed for each group. e Mosaicism of the germline of F0 founders for cx30.3 and cx34.4 KI was determined by the percentage of F1 carrying the KI cassette. PCR-positive cx30.3+/+eGFP or cx34.4+/+eGFP F1 were shown in blue and PCR-negative cx30.3+/+ or cx34.4+/+ F1 were in orange. More than 70 F1 were examined for each PCR-positive F0 founder.
|