- Title
-
Myeloid cells interact with a subset of thyrocytes to promote their migration and follicle formation through NF-κB
- Authors
- Yang, R.M., Song, S.Y., Wu, F.Y., Yang, R.F., Shen, Y.T., Tu, P.H., Wang, Z., Zhang, J.X., Cheng, F., Gao, G.Q., Liang, J., Guo, M.M., Yang, L., Zhou, Y., Zhao, S.X., Zhan, M., Song, H.D.
- Source
- Full text @ Nat. Commun.
|
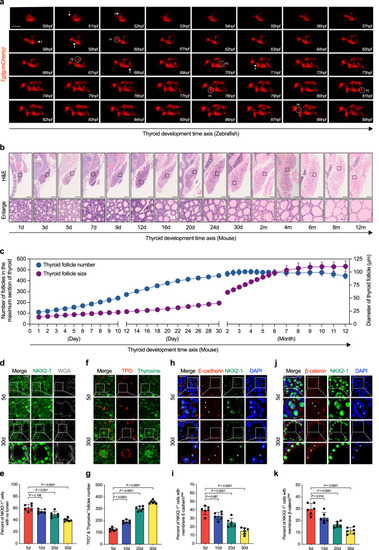
Gradual transformation of solid primordium into arborized follicles in mice and zebrafish. |
|
Identification of motile thyrocytes by scRNA-seq from the developing mice thyroid tissues. |
|
NF-κB is activated in the central immature thyrocytes of mice by spatial transcriptome sequencing. |
|
NF-κB signaling is activated in TFC with higher migratory capacity in zebrafish and mice. |
|
NF-κB inhibition affect thyroid follicle formation in mice. |
|
Myeloid cells secrete Tnf-α to promote NF-κB activation in mice thyroid epithelial cells. |
|
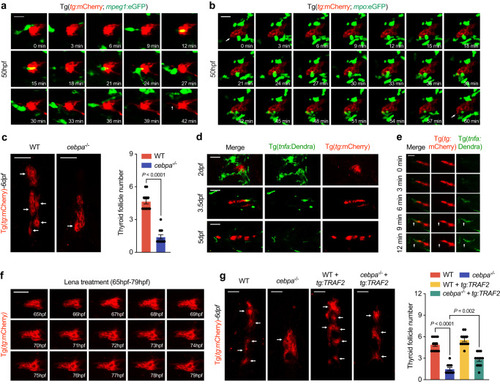
Cells of myeloid lineage secrete Tnf-α to promote NF-κB activation in zebrafish thyroid epithelial cells. |