- Title
-
Generation and validation of a myoglobin knockout zebrafish model
- Authors
- Hejlesen, R., Kjær-Sørensen, K., Fago, A., Oxvig, C.
- Source
- Full text @ Transgenic. Res.
|
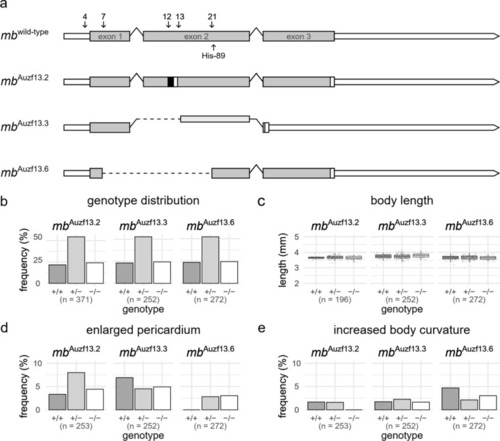
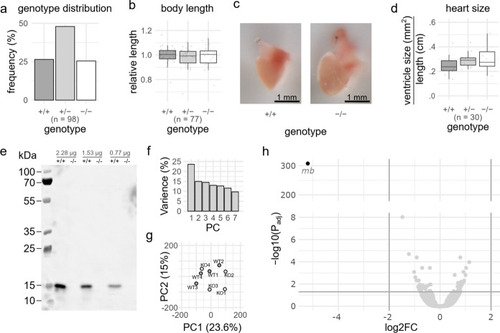
Generated myoglobin ( PHENOTYPE:
|
|
Characterization of adult (3–6 months post fertilization) |