- Title
-
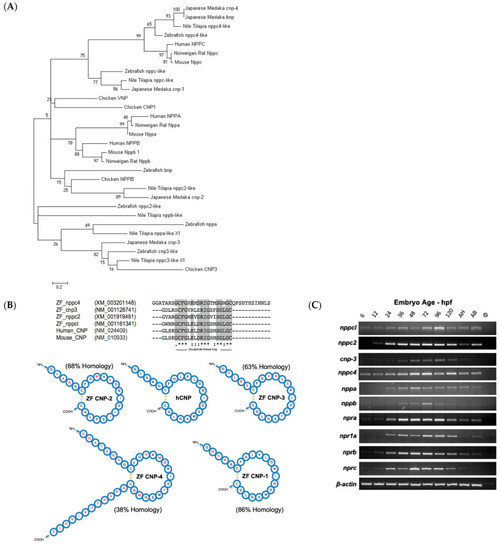
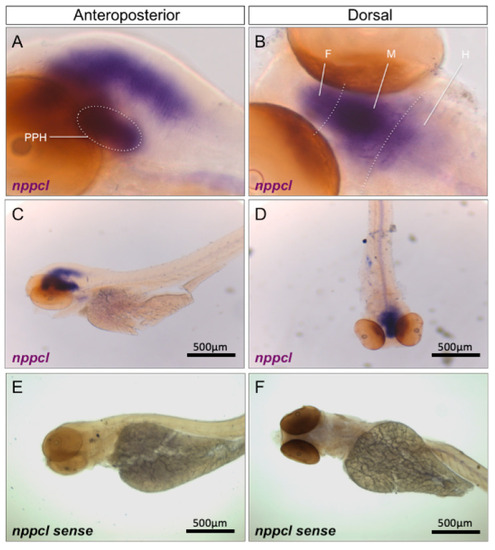
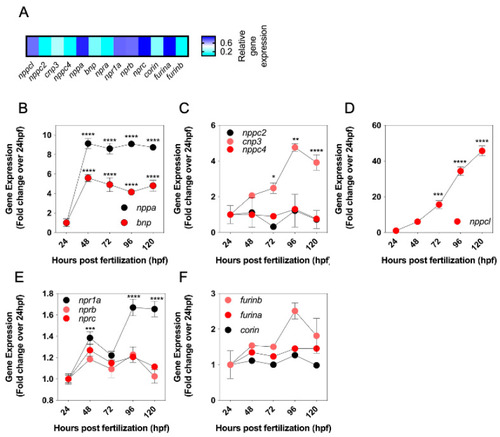
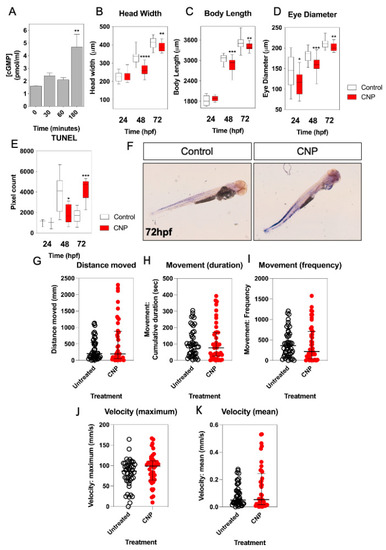
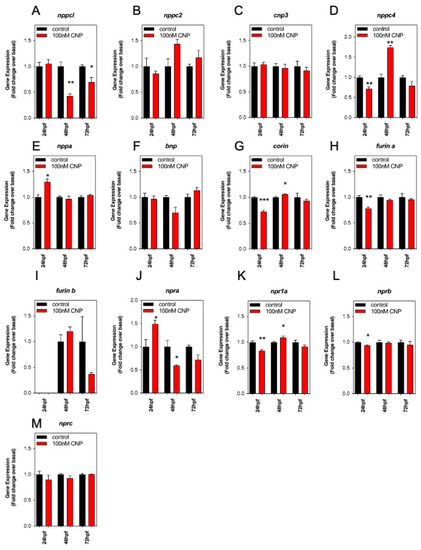
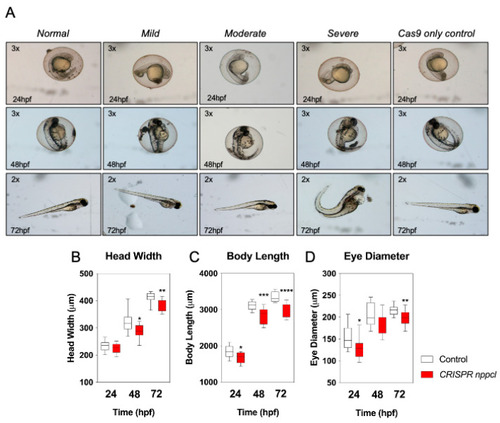
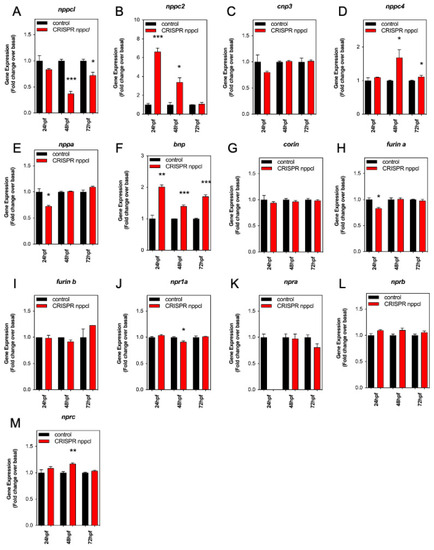
Pharmacological and Genetic Disruption of C-Type Natriuretic Peptide (nppcl) Expression in Zebrafish (Danio rerio) Causes Stunted Growth during Development
- Authors
- Lessey, A.J., Mirczuk, S.M., Chand, A.N., Kurrasch, D.M., Korbonits, M., Niessen, S.J.M., McArdle, C.A., McGonnell, I.M., Fowkes, R.C.
- Source
- Full text @ Int. J. Mol. Sci.
|
( |
|
( |
|
( |
|
( |
|
( |
|
( |
|
( |