- Title
-
CORRECTION: rbpms2 functions in Balbiani body architecture and ovary fate
- Authors
- Kaufman, O.H., Lee, K., Martin, M., Rothhämel, S., Marlow, F.L.
- Source
- Full text @ PLoS Genet.
|
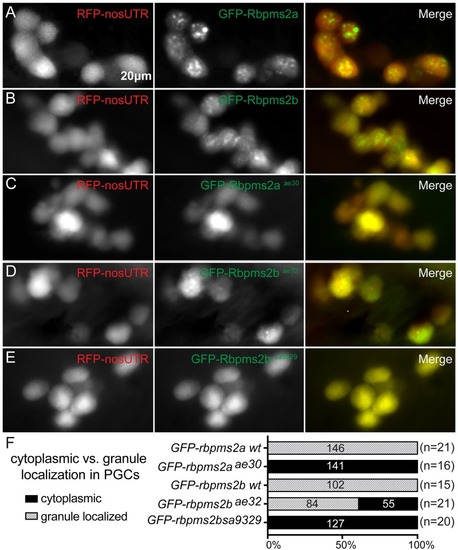
(A-B) Wild-type GFP-Rbpms2a/b localize to germ granules of PGCs. PGCs are labeled with RFP- |
|
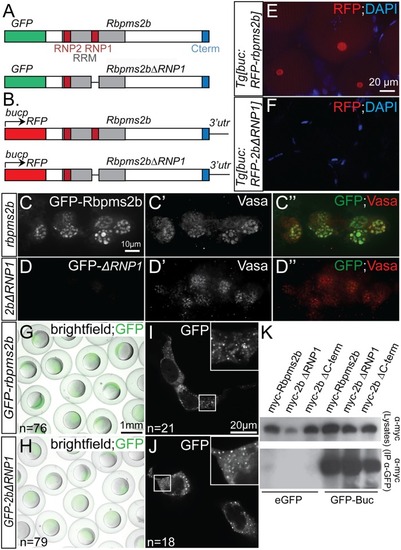
(A) Schematic representing constructs used in granule localization assays. (B) Schematic representing transgenic constructs used to express Rbpms2b in oocytes. (C-C”) Wild-type GFP-Rbpms2b localizes with Vasa in germ granules of PGCs. (D-D”) GFP-Rbpms2bΔRNP1, a deletion of 7 residues within the RRM domain, does not localize to germ granules. (E-F) RFP-Rbpms2b expressed in transgenic Tg[buc:RFP-rbpms2b] ovaries localizes to the Bb (E), while RFP-Rbpms2bΔRNP1 does not (F). (G-J) GFP-Rbpms2b and GFP-Rbpms2bΔRNP1 are stably expressed in gastrula stage embryos (G,H), as well as the granules of HEK293 cells (I, J). (K) GFP immunoprecipitation of Myc- tagged Rbpms2b, Rbpms2bΔRNP1, and Rbpms2bΔC-terminus, a deletion of the last 7 residues within the conserved C-term, demonstrates that the RNP1 domain or conserved C-terminus is not required for interaction with GFP-Bucky ball. |
|
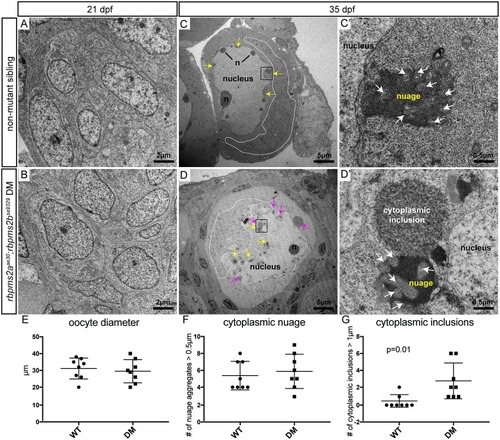
(A-B) Comparable clusters of gonocytes from d21 non-mutant siblings (A), and |