- Title
-
The neurodevelopmental gene MSANTD2 belongs to a gene family formed by recurrent molecular domestication of Harbinger transposons at the base of vertebrates
- Authors
- Etchegaray, E., Baas, D., Naville, M., Haftek-Terreau, Z., Volff, J.N.
- Source
- Full text @ Mol Bio Evol
|
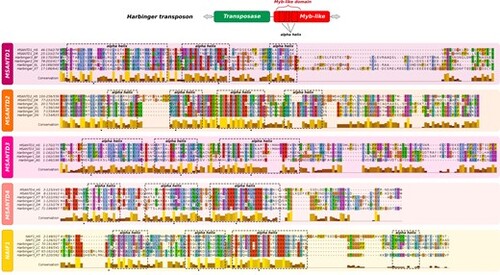
Multiple alignments of the Myb-like domain of MSANTD proteins and their closest Myb-like proteins from Harbinger transposons. Predicted alpha-helix motifs are represented with dashed squares; bulky aromatic residues, which are essential for alpha helix structure stabilization, are indicated by black stars. The conservation score represented for each residue is measured considering both residue identities and conservation of physico-chemical classes. AG, Anopheles gambiae; DR, Danio rerio; EL, Esox lucius; GA, Gasterosteus aculeatus; GG, Gallus gallus; HS, Homo sapiens; LC, Latimeria chalumnae; OL, Oryzias latipes; ON, Oreochromis niloticus; SS, Salmon salar; TF, Takifugu flavidus; XT, Xenopus tropicalis. |
|
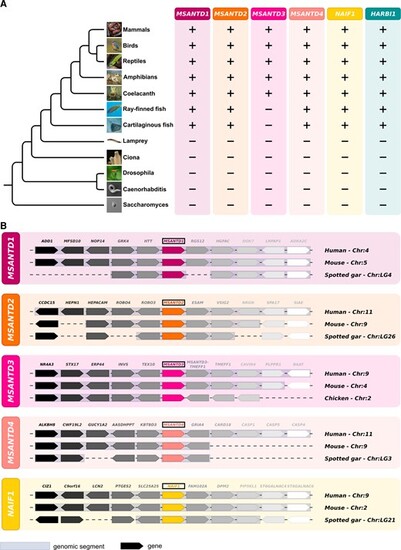
(A) Phylogenetic distribution of Harbinger-derived genes and Harbinger transposons. Presence (+) or absence (–) of these genes in the different lineages is indicated. (B) Synteny analysis of Myb-like-derived genes between human, mouse and spotted gar (nonteleost ray-finned fish) or chicken (for MSANTD3, which is absent from both cartilaginous and ray-finned fish). Species names and genomic locations are shown on the right. For each gene the same color stands for orthologous genes. |
|
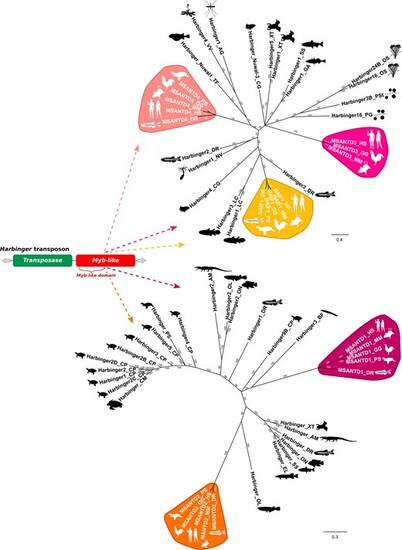
Phylogenetic relationships between MSANTD proteins and their closest Myb-like proteins from Harbinger transposons. Trees were constructed using the Bayesian method (Huelsenbeck and Ronquist 2001). Only branch support values higher than 50% are shown. AG, Anopheles gambiae; AM, Alligator mississippiensis; BF, Branchiostoma floridae; CG, Crassostrea gigas; CM, Chelonia mydas; CP, Chrysemys picta; DR, Danio rerio; EL, Esox lucius; GA, Gasterosteus aculeatus; GG, Gallus gallus; HS, Homo sapiens; LC, Latimeria chalumnae; MM, Mus musculus; NV, Nematostella vectensis; OL, Oryzias latipes; ON, Oreochromis niloticus; OS, Oryza sativa; PG, Puccinia graminis; PS, Pelodiscus sinensis; PSt, Puccinia striiformis; SS, Salmon salar; TF, Takifugu flavidus; VV, Vitis vinifera; XT, Xenopus tropicalis). Silhouette images from phylopic.org. |
|
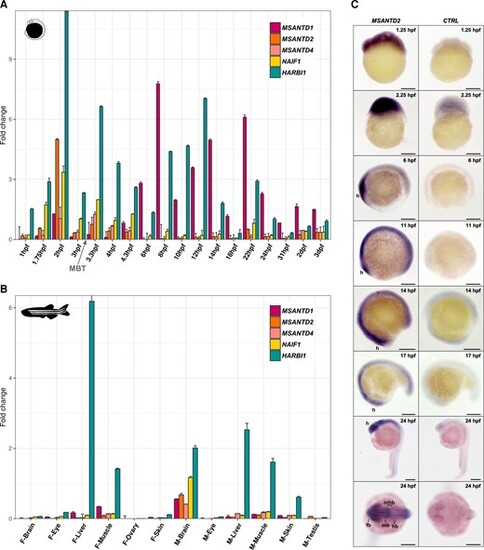
Expression of Harbinger-derived genes in zebrafish embryos and adults. (A) Relative gene expression determined by qPCR compared to the 18S housekeeping gene during embryonic development (from 1hpf to 3dpf). The MBT is shown with an arrow. (B) Relative gene expression determined by qPCR compared to the 18S housekeeping gene in female (F) and male (M) adult tissues. (C) Whole-mount in situ hybridization of MSANTD2 in zebrafish embryos from 1.25hpf to 24hpf using a sense probe as a control. The head or future head region is indicated with ‘h’. fb, forebrain; mb, midbrain; hb, hindbrain. Scale bars: 200 μm. |
|
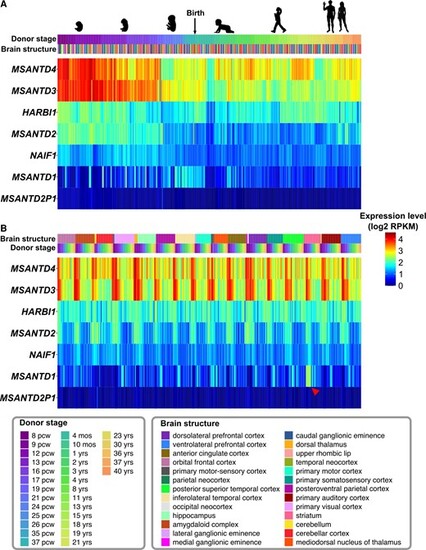
Expression of Harbinger-derived genes in human brain before and after birth according to donor stages (A) or brain structures (B). For each gene, the expression is shown in log2 reads per kilobase per million (RPKM) for different donor stages (pcw, postconceptional weeks; mos, months; yrs, years) and in different brain structures, represented with multiple colors. Data were obtained from the BrainSpan Atlas (www.brainspan.org) (Miller et al. 2014). The striatum-specific expression of MSANTD1 is indicated with a red arrowhead. Silhouette images are from lifesizesilhouette.com and (Haniffa et al. 2021). |
|
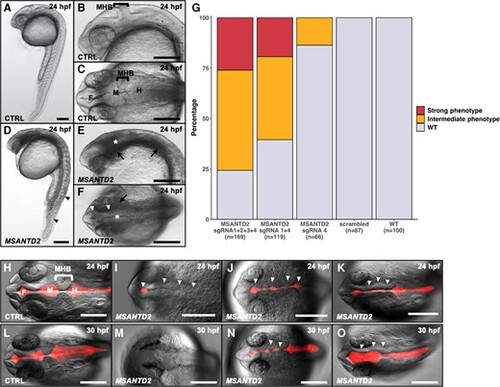
MSANTD2 CRISPR/Cas9 embryo phenotypes. Embryos injected with control (A–C, CTRL) or four MSANTD2-directed sgRNAs (D–F, MSANTD2). A/D and B/E present lateral views of whole embryos and of the head region, respectively. C/F show dorsal views of the embryo head region. Embryos injected with four sgRNAs showed developmental delays as well as tail and nervous system malformations compared to control embryos at 24hpf (A–F). Tail curvation and not well-defined somites are shown with black arrowheads (D). Default in neural tube folding and cell aggregates around the nervous system are indicated with white arrowheads and black arrows, respectively (E–F). Not well-formed MHB are shown with white stars (*) (E–F). (G) Proportions of F0 zebrafish phenotypes. Embryos were injected with sgRNA 1 + 2 + 3 + 4, sgRNA 1 + 4 and sgRNA 4 targeting MSANTD2, four scrambled sgRNAs or noninjected (WT), and were scored for phenotypes at 24hpf. Percentages with strong (red), intermediate (orange) or no phenotypes (gray–WT) are shown. Strong phenotypes: developmental delays, tail malformations, nervous system malformations, aggregates; intermediate phenotypes: developmental delays, nervous system malformations, no or few aggregates, no tail malformation. (H–O): Dextran Texas Red injection in brain ventricles of 24hpf (H–K) or 30hpf (L–O) embryos injected with control (H, L, CTRL) or four MSANTD2-directed sgRNAs (I–K, M–O, MSANTD2) (dorsal view). At each stage, three pictures of MSANTD2 CRISPR/Cas9 embryos were compared to a control picture in order to represent phenotype variability. The proportion of embryos presenting the same phenotypes are the following: I—31%, J—47%, K—22% at 24hpf and M—14%, N—43%, O—43% at 30hpf. All cases illustrate neural tubes misfolding (shown with white arrowheads) particularly in the MHB region. We also observed smaller red fluorescent areas in the forebrain and midbrain regions, indicating reduction of these ventricles. F, forebrain, M, midbrain; H, hindbrain. Scale bar 200 µm. |
|
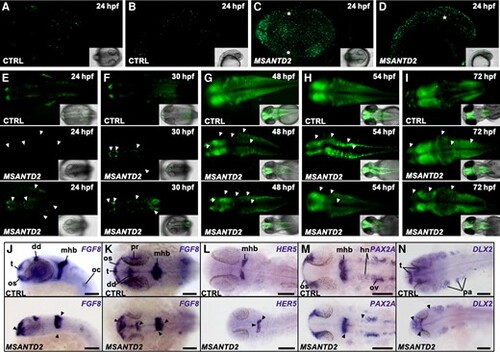
Effects of MSANTD2 inactivation by CRISPR/Cas9 in zebrafish embryos. (A–D) Acridine orange staining of embryos injected with control (CTRL) or four MSANTD2-directed sgRNAs (MSANTD2) at 24hpf. Numerous dead cells were visible in MSANTD2 CRISPR/Cas9 embryos compared to control embryos. Regions of cell aggregates were indicated with white stars. (E–I) Embryos injected with control (CTRL) or four MSANTD2-directed sgRNAs (MSANTD2) in Tg(elavl3:GCaMP3) zebrafish embryos. At each stage, two pictures of MSANTD2 CRISPR/Cas9 embryos were compared to a control in order to represent the variability of impaired neuronal pattern. Differentiated neurons were marked by green fluorescence. The results showed general anatomical defects characterized by different patterns of fluorescence between MSANTD2-mutated and control embryos (shown with white arrowheads). Abnormal pattern of neuronal marking was not only explained by developmental delay, since MSANTD2 mutated embryos are still different from control embryos at later stages. (J–N): Expression of FGF8 (J, K), HER5 (L), PAX2A (M) and DLX2 (N) at 24hpf in embryos injected with control (CTRL, top) or four MSANTD2-directed sgRNAs (MSANTD2, bottom). J present lateral and K/L/M/N dorsal views of the head region of the embryos, respectively. Differences in gene expression patterns between MSANTD2-mutated and control embryos are indicated with black arrowheads. Dd, dorsal diencephalon; hn, hindbrain neurons; oc, otic capsule; os, optic stalks; ov, otic vesicle; pa, pharyngeal arches; pr, proximal part of retina; t, telencephalon; tp, thyroid primordium. For each picture at least n = 10 embryos were observed. |