- Title
-
Hypoxia-induced miR-210-3p expression in lung adenocarcinoma potentiates tumor development by regulating CCL2-mediated monocyte infiltration
- Authors
- Arora, L., Patra, D., Roy, S., Nanda, S., Singh, N., Verma, A.K., Chakraborti, A., Dasgupta, S., Pal, D.
- Source
- Full text @ Mol. Oncol.
|
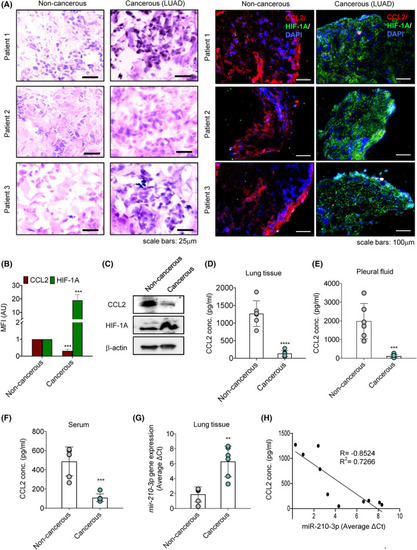
Hypoxic tumor microenvironment alters the expression of CCL2 and |
|
Hypoxia‐induced miR‐210‐3p directly inhibits |
|
HIF‐1A regulates miR‐210‐3p mediated |
|
Hypoxic tumor microenvironment in 3D lung adenocarcinoma spheroids regulates CCL2 mediated monocyte infiltration and promotes TAM polarization (A,B) Representative phase‐contrast (A), and Calcein‐AM fluorescence staining (B) images of 3D spheroids of BEAS‐2B, A549, and NCI‐H460 cells on day 10 showing THP‐1 monocytes infiltrations in normoxic and hypoxic conditions for 24 h. Scale bars: 100 μm. (C,D) Flow cytometric analysis of CD11b level (C) and its quantifications (D) indicated THP‐1 monocytes infiltration in 3D spheroids of BEAS‐2B or A549 or NCI‐H460 cells incubated in normoxic and hypoxic conditions for 24 h. (E,F) RT‐qPCR analysis showing the relative abundance of |
|
Anti‐miR‐210‐3p LNA delivery increases macrophage burden promoting tumor regression in the A549 lung cancer xenograft zebrafish model. (A) Schematic diagram representing experimental design indicating days of zebrafish A549 tumor xenograft model development followed by the administration of anti‐miR‐210‐3p LNA and collection of tumor tissue and blood samples from zebrafish. (B) Representative images of A549 tumor xenograft zebrafish (day 14/control) treated without or with anti‐miR‐210‐3p LNA for indicated timepoints (left panel) ( |
|
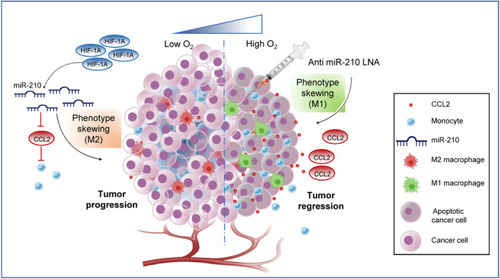
Model representing miR‐210‐3p as a potential therapeutic target for lung adenocarcinoma. Left panel: LUAD solid tumor exhibits hypoxia‐associated stabilization of HIF‐1A, which by promoting miR‐210‐3p level directly involved in downregulation of CCL2 and macrophage infiltration and also favors macrophage TAM phenotype supporting tumor progression. Right panel: Therapeutic intervention of anti‐miR210‐3p LNA augment CCL2 mediated monocyte infiltration and skewing towards macrophage antitumoral M1 phenotype leading to tumor regression. |