- Title
-
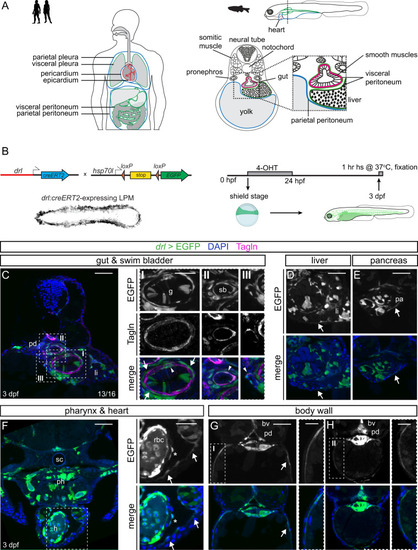
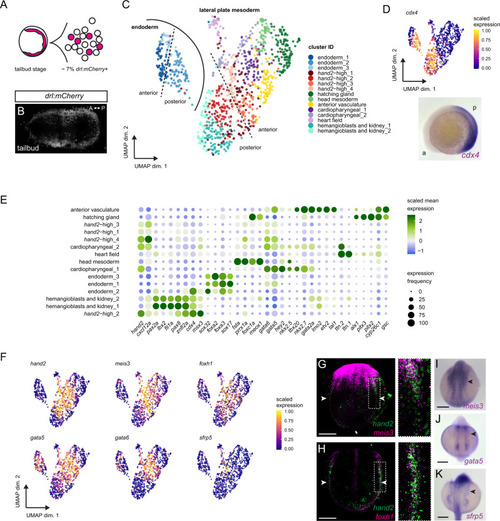
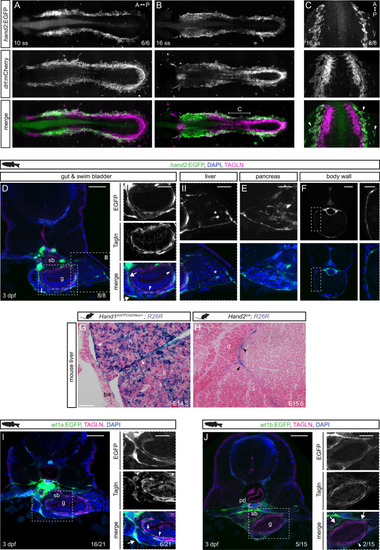
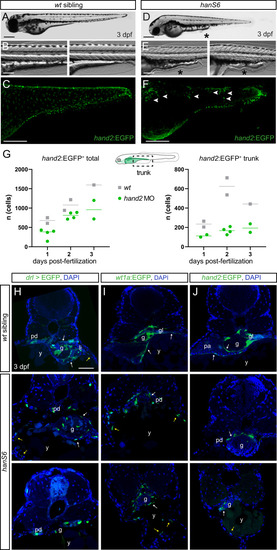
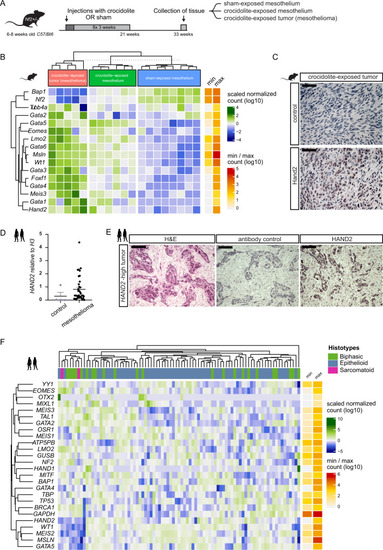
Hand2 delineates mesothelium progenitors and is reactivated in mesothelioma
- Authors
- Prummel, K.D., Crowell, H.L., Nieuwenhuize, S., Brombacher, E.C., Daetwyler, S., Soneson, C., Kresoja-Rakic, J., Kocere, A., Ronner, M., Ernst, A., Labbaf, Z., Clouthier, D.E., Firulli, A.B., Sánchez-Iranzo, H., Naganathan, S.R., O'Rourke, R., Raz, E., Mercader, N., Burger, A., Felley-Bosco, E., Huisken, J., Robinson, M.D., Mosimann, C.
- Source
- Full text @ Nat. Commun.
|
|
|
|
|
|
|
|
|
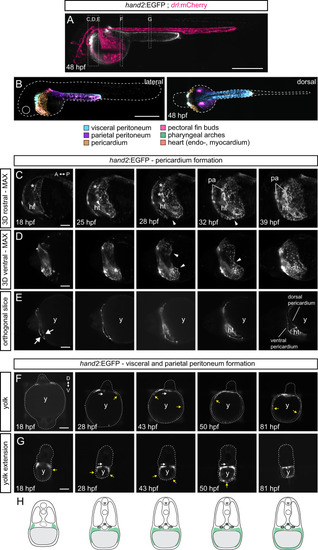
Multiday, multi-angle SPIM of |
|
|
|
|