|
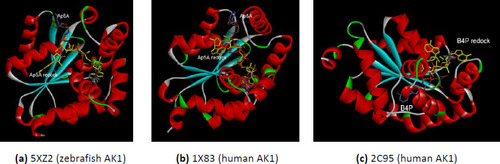
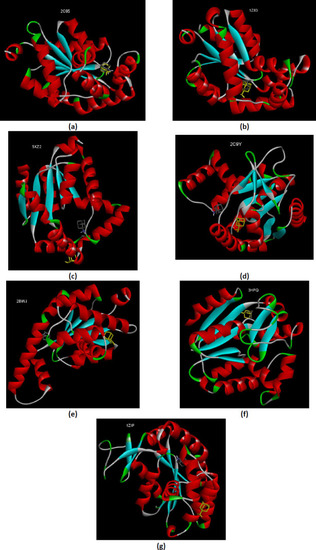
The comparative interactions of the AK1 isoforms with the analog substrates (Ap5A or B4P) after redocking. The redocked conformations are shown in yellow. (a) zebrafish AK1 (PDB ID 5XZ2); (b) human AK1 (PDB ID 1X83); (c) human AK1 (PDB ID 2C95). |
|
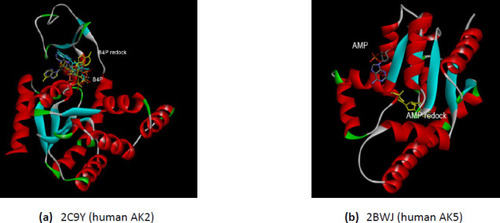
(a) The interactions comparative of the human AK2 (PDB ID 2C9Y) with the B4P after redocking; (b) The comparative interactions of the human AK5 (PDB ID 2BWJ) with the AMP after redocking. The redocked conformations are shown in yellow. |
|
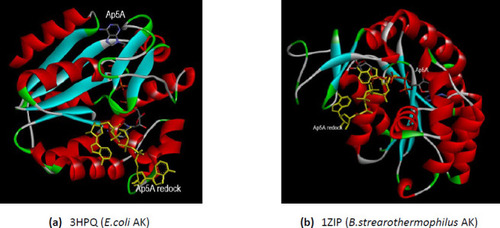
The comparative interactions of the bacterial AKs with the analog substrate Ap5A after redocking. The redocked conformations are shown in yellow. (a) E.coli AK (PDB ID 3HPQ); (b) B.stearothermophilus AK (PDB ID 1ZIP). |
|
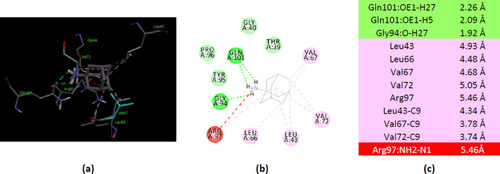
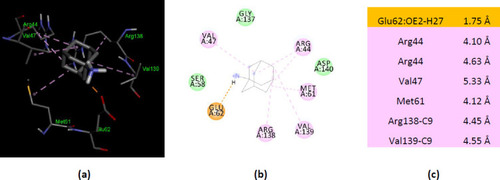
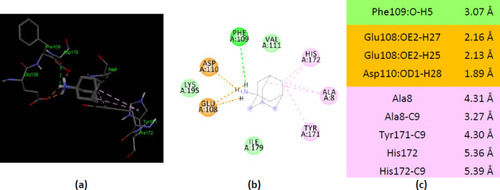
Interactions of ionized amantadine with human AK1, PDB ID 1Z83. (a) 3D display of ionized amantadine interaction as ligand with the 1Z83 residues; (b) code color for interactions: in green are shown conventional hydrogen bonds, light green – van der Waals forces, mauve – alkyl hydrophobic interactions, red – unfavorable positive-positive interactions; (c) the distances (Å) of the 1Z83 - ionized amantadine interactions. |
|
Interactions of ionized amantadine with human AK1, PDB ID 2C95. ( |
|
Interactions of ionized amantadine with zebrafish AK1, PDB ID 5XZ2. (a) 3D display of ionized amantadine interaction as ligand with the 5XZ2 residues; (b) code color for interactions: in green are shown conventional hydrogen bonds, orange – salt bridge, light green – van der Waals forces, mauve – alkyl hydrophobic interactions; (c) the distances (Å) of the 5XZ2 - ionized amantadine interactions. |
|
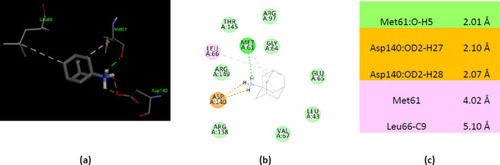
Interactions of ionized amantadine with human AK2, PDB ID 2C9Y. (a) 3D display of ionized amantadine interaction as ligand with the 2C9Y residues; (b) code color for interactions: in green are shown conventional hydrogen bonds, orange – salt bridge, light green – van der Waals forces, mauve – alkyl hydrophobic interactions; (c) the distances (Å) of the 2C9Y-ionized amantadine interactions. |
|
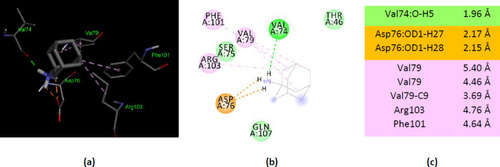
Interactions of amantadine with human AK5, PDB ID 2BWJ. (a) 3D display of ionized amantadine interaction as ligand with the 2BWJ residues; (b) code color for interactions: in orange are shown salt bridge, light green – van der Waals forces, mauve – alkyl hydrophobic interactions, red – unfavorable donor-donor interactions; (c) the distances (Å) of the 2BWJ - ionized amantadine interactions. |
|
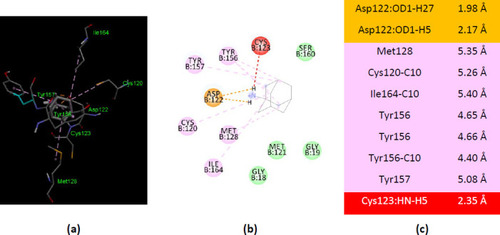
Interactions of ionized amantadine with AK from E.coli, PDB ID 3HPQ. (a) 3D display of amantadine interaction as ligand with the 3HPQ residues; (b) code color for interactions: in green are shown conventional hydrogen bonds, orange – salt bridge, light green – van der Waals forces, mauve – alkyl hydrophobic interactions; (c) the distances (Å) of the 3HPQ - ionized amantadine interactions. |
|
Interactions of ionized amantadine with AK from B.stearothermophilus, PDB ID 1ZIP. (a) 3D display of amantadine interaction as ligand with the 1ZIP residues; (b) code color for interactions: in green are shown conventional hydrogen bonds, orange – salt bridge, light green – van der Waals forces, mauve – alkyl hydrophobic interactions; (c) the distances (Å) of the 1ZIP - ionized amantadine interactions. |
|
Interactions of unionized and ionized amantadine with AKs. (a) human AK1 (PDB ID 2C95); (b) human AK1 (PDB ID 1Z85) (c) zebrafish AK1 (PDB ID 5XZ2); (d) human AK2 (PDB ID 2C9Y); (e) human AK5 (PDB ID 2BWJ) (f) AK from E.coli (PDB ID 3HPQ); (g) AK from B.stearothermophilus (PDB ID 1ZIP); unionized amantadine is shown in yellow. |
|
Multiple sequence alignment of the AKs sequences. In gray background are marked the residues belong to Walker A motif – phosphate-binding loop or P-loop; in yellow background is marked the NMP binding region and the residues that belong to AMP binding region are italic-bolded; in blue background is marked the LID region; with red are marked the residues (van der Waals, conventional hydrogen bond or alkyl interactions) which interact with amantadine ionized form (with –NH3+ group); the residues which interact with amantadine unionized form (with –NH2 group) are bolded-underlined; the residues which interact with amantadine unionized and ionized are red and bolded-underlined; AKEC is AK from E.coli; AKBS is AK from B.stearothermophilus; with dot “.” are marked the semi-conservative replacements; with colon “:” are marked the conservative replacements; with “*” are marked the identities of the residues. |
|
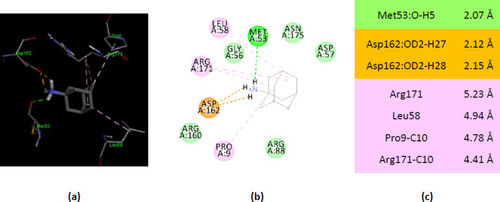
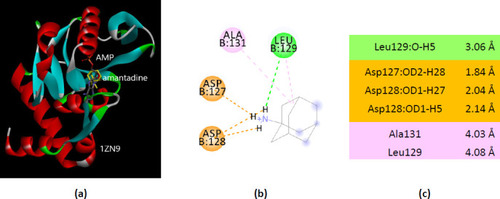
Interactions of ionized amantadine with APRT co-crystallized with AMP, PDB ID 1ZN9. ( |
|
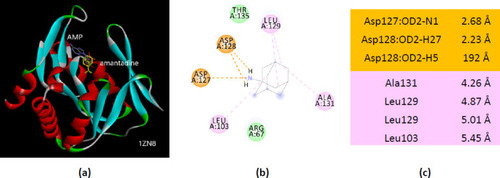
Interactions of ionized amantadine with APRT co-crystallized with AMP, PDB ID 1ZN8. (a) 3D display of the amantadine (shows in yellow) and AMP in binding pocket; (b) code color for interactions: in orange are shown the salt bridges, light green – van der Waals forces, mauve – alkyl hydrophobic interactions; (c) the distances (Å) of the 1ZN8 - ionized amantadine interactions. |
|
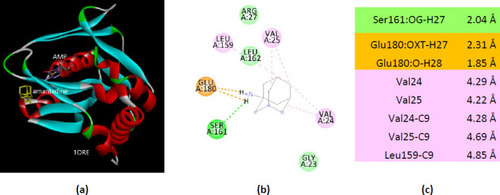
Interactions of ionized amantadine with APRT co-crystallized with AMP, PDB ID 1ORE. (a) 3D display of the amantadine (shows in yellow) and AMP in binding pocket; (b) code color for interactions: in orange are shown the salt bridges, light green – van der Waals forces, mauve – alkyl hydrophobic interactions; (c) the distances (Å) of the 1ORE - ionized amantadine interactions. |
|
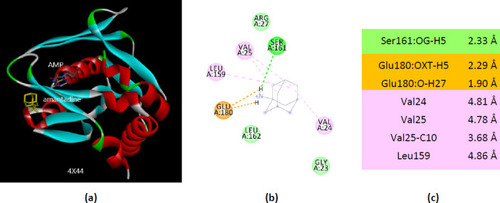
Interactions of ionized amantadine with APRT co-crystallized with AMP, PDB ID 4X44. (a) 3D display of the amantadine (shows in yellow) and AMP in binding pocket; (b) code color for interactions: in orange are shown the salt bridges, light green – van der Waals forces, mauve – alkyl hydrophobic interactions; (c) the distances (Å) of the 4X44 - ionized amantadine interactions. |
|
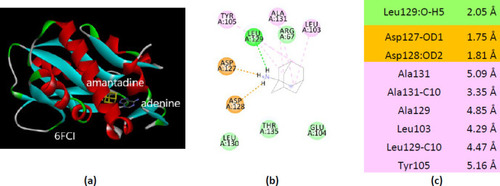
Interactions of ionized amantadine with APRT co-crystallized with adenine, PDB ID 6FCI. (a) 3D display of the amantadine (shows in yellow) and adenine in binding pocket; (b) code color for interactions: in orange are shown the salt bridges, light green – van der Waals forces, mauve – alkyl hydrophobic interactions; (c) the distances (Å) of the 6FCI - ionized amantadine interactions. |
|
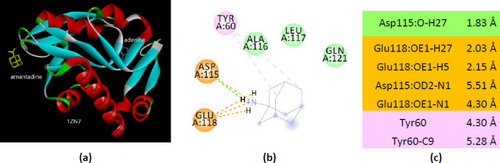
Interactions of ionized amantadine with APRT co-crystallized with adenine, PDB ID 1ZN7. (a) 3D display of the amantadine (shows in yellow) and adenine in binding pocket; (b) code color for interactions: in orange are shown the salt bridges, light green – van der Waals forces, mauve – alkyl hydrophobic interactions; (c) the distances (Å) of the 1ZN7 - ionized amantadine interactions. |
|
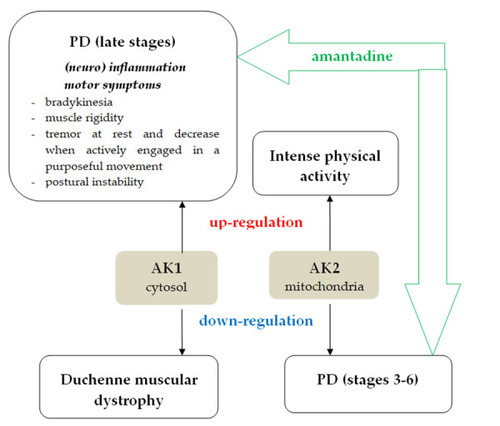
The involvement of human AK1 and human AK2 in PD, inflammation, and muscle activity. The amantadine treatment efficacy in PD developed the hypothesis that amantadine – AK interactions. |