- Title
-
Genetic dissection of novel myopathy models reveals a role of CapZα and Leiomodin 3 during myofibril elongation
- Authors
- Berger, J., Berger, S., Mok, Y.S.G., Li, M., Tarakci, H., Currie, P.D.
- Source
- Full text @ PLoS Genet.
|
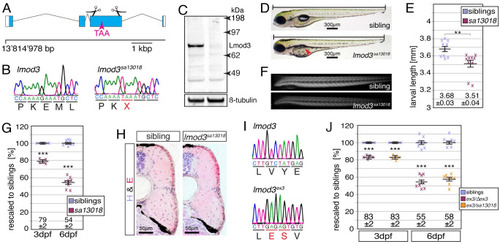
(A) The mutant line EXPRESSION / LABELING:
PHENOTYPE:
|
|
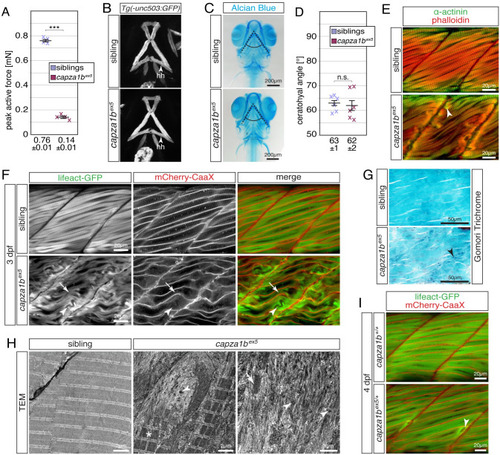
(A) Peak active force of single-twitch contractions generated by 6-dpf-old individual |
|
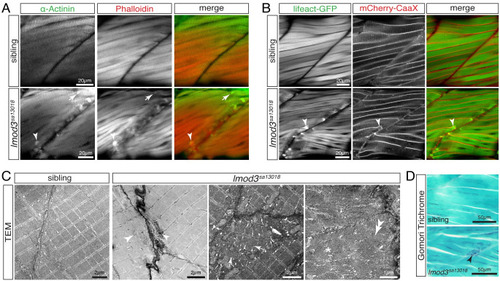
(A) Immunohistochemistry using antibodies against α-Actinin (green) and phalloidin staining (red) marked aggregates located close to vertical myosepta of 4-dpf-old |
|
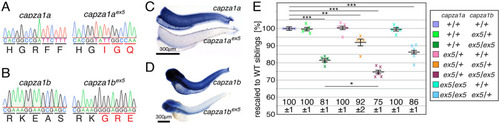
(A) EXPRESSION / LABELING:
PHENOTYPE:
|
|
(A) Whereas 6-dpf-old siblings were able to generate a peak active force of 0.76 ± 0.01 mN, the force generated by EXPRESSION / LABELING:
PHENOTYPE:
|
|
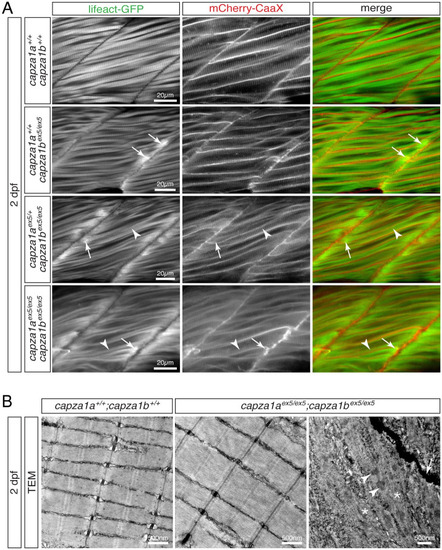
(A) At 2 dpf, Lifeact-GFP (green) and mCherry-CaaX (red) highlighted the sarcomere organisation and myofibril striation within muscle fibres of WT siblings. In two out of four analysed |
|
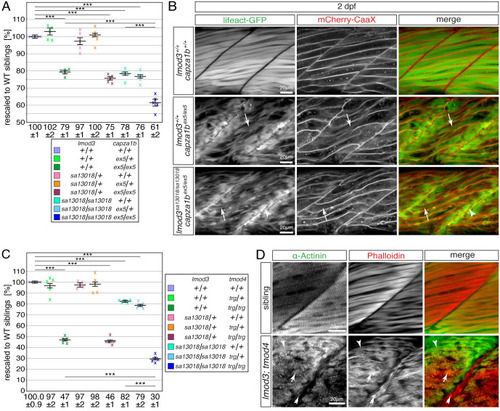
(A) After rescaling to WT siblings (100 ± 1%), the significant reduction of birefringence of single EXPRESSION / LABELING:
PHENOTYPE:
|