- Title
-
Rhythmic Clock Gene Expression in Atlantic Salmon Parr Brain
- Authors
- Bolton, C.M., Bekaert, M., Eilertsen, M., Helvik, J.V., Migaud, H.
- Source
- Full text @ Front. Physiol.
|
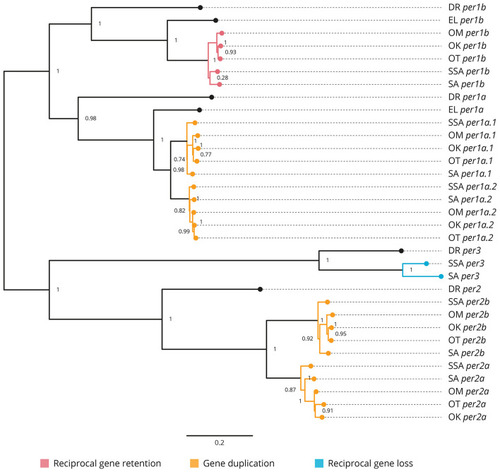
Phylogenetic relationship of the period clock gene family (per). Gene family members from Danio rerio (DR), Esox Lucius (EL), Salmo salar (SSA), Salvelinus alpinus (SA), Oncorhynchus mykiss (OM), Oncorhynchus kisutch (OK), and Oncorhynchus tshawytscha (OT) were re-annotated. Values on the relevant node depict the bootstrap values. Reciprocal gene retention (red), gene duplication (orange), and reciprocal gene loss (blue). |
|
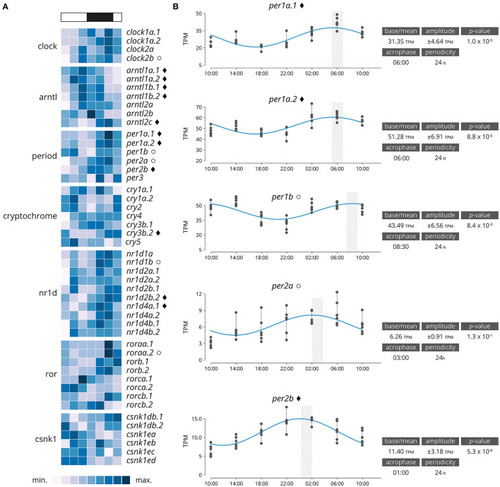
(A) Heatmap displaying average diel expression of identified clock genes under constant LD (12:12, n = 6 per time point). The heatmap of the relative expression of each individual gene [scaled from lowest expression to highest expression]. Black diamond indicates significantly cyclic gene (p < 0.001) [JTK and RAIN analysis], White circles denote rhythmic genes (p < 0.001) [RAIN analysis]. (B) Significantly clock gene expression. Parameters of the cyclic sin-cosine function calculated by MetaCycle with JTK for diel expression of clock genes in brains collected from Atlantic salmon smolt exposed to an LD 12:12 photoperiod. |
|
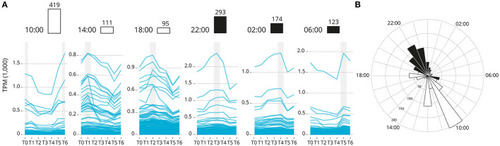
Genes groups based on their acrophase (expression peak) synchronicity. (A) Based on the average recorded TPM values. The number of gene is reported over the barplot (white and black boxes denote light or darkness conditions, respectively). Sub-figures exclude three highly expressed genes that bias the scale of the graphic: Ependymin-1 (> 10,000 TPM) Ependymin-2 (> 5,000 TPM) and calcium voltage-gated channel auxiliary subunit alpha 2 delta 4 (> 2,000 TPM). (B) Rose plot of the phase distribution of highly significantly circadian gene expression (p < 0.001, JTK analysis). Color coding indicates whether the phase occurs during the light (white) or dark (black) (12:12 LD). |