- Title
-
A Comparative Analysis of Gene and Protein Expression Throughout a Full 28-Day Retinal Regeneration Time-Course in Adult Zebrafish
- Authors
- Kramer, A.C., Gurdziel, K., Thummel, R.
- Source
- Full text @ Front Cell Dev Biol
|
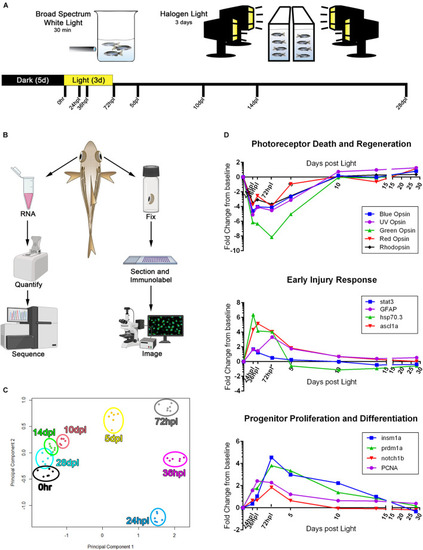
3′mRNA-seq analysis following phototoxic lesion and regeneration confirms gene expression trends known in the field. |
|
Green and red cone photoreceptor morphology paired with gene expression of isoforms throughout a 28 day lesion and regeneration time-course. |
|
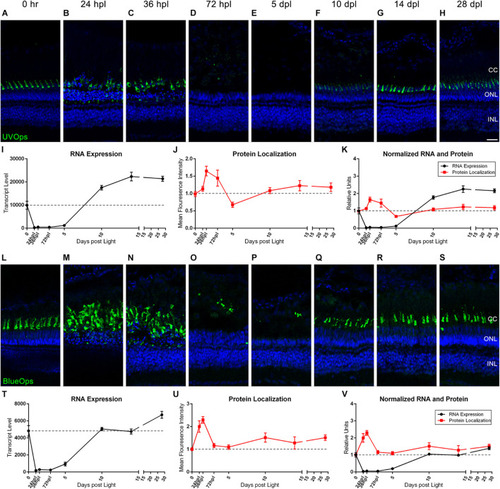
UV and Blue cone photoreceptor morphology paired with gene expression throughout a 28 day lesion and regeneration time-course. |
|
Rod photoreceptors degenerate in coordination with the infiltration of 4C4+ microglia/macrophages. |
|
The Müller glia response to phototoxic lesion paired with gene expression signatures. (A–H) The Müller glia (MG) response to phototoxic lesion is demonstrated in these retinal sections collected at baseline (0 h) through 28 days post phototoxic lesion (dpl), hours post light are denoted (hpl) by immunolabeling the intermediate filaments of MG with anti-GFAP (green) and their entry into the cell cycle with PCNA (red). Nuclei were stained blue with TO-PRO-3. Gliosis is highlighted in a biphasic manner: in the ONL MG end-feet at 24 hpl, and then spanning the length of MG at 5 dpl. The PCNA localization demonstrates single MG nuclei entering the cell cycle at 36 hpl with peak proliferation of MGPCs at 72 hpl (n = 5–6). (I) Graph of transcript pseudo-counts for GFAP from 3′mRNA-seq of individual adult zebrafish retinas for each timepoint (n = 6). (J) ImageJ pixel intensity quantification for the GFAP signal in the confocal images normalized to 1, demonstrating relative intensity of protein localization within the retina. (K) Overlay of GFAP RNA expression normalized to 1 and ImageJ protein localization normalized to one. The discordance in the gene expression and protein localization suggests that the first GFAP protein intensity peak at 24 hpl is due to a redistribution of existing GFAP protein and the second protein intensity peak at 5 dpl is due to increased transcription of GFAP. (L) Graph of transcript pseudo-counts for PCNA from 3′mRNA-seq of individual adult zebrafish retinas for each timepoint (n = 6). (M) ImageJ pixel intensity quantification for the PCNA signal in the confocal images normalized to 1, demonstrating relative intensity of protein localization within the retina. (N) Overlay of PCNA RNA expression normalized to 1 and ImageJ protein localization normalized to one. Scale bar represents 5 μm. |
|
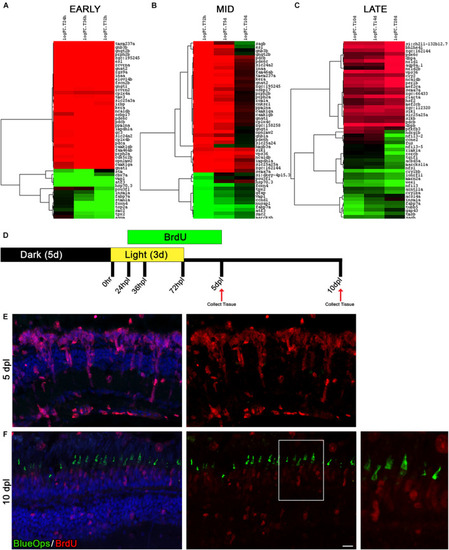
The time window between days 5 and 10 of regeneration represents a distinct turning point toward differentiation. |
|
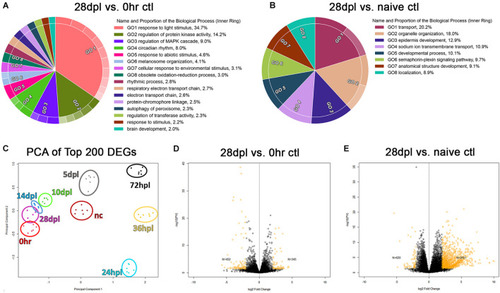
28 days post light does not represent transcriptional recovery to either the 0 h, or naïve control transcriptional baseline. |
|
Common genes studied in retinal regeneration with dark adapted 0 h control, and naïve, non-dark adapted control baselines. All graphs in this figure represent transcript pseudo-counts for each of the genes listed above the plots from 3′mRNA-seq of individual adult zebrafish retinas for each timepoint ( |