- Title
-
Meiotic Chromosome Dynamics in Zebrafish
- Authors
- Imai, Y., Olaya, I., Sakai, N., Burgess, S.M.
- Source
- Full text @ Front Cell Dev Biol
|
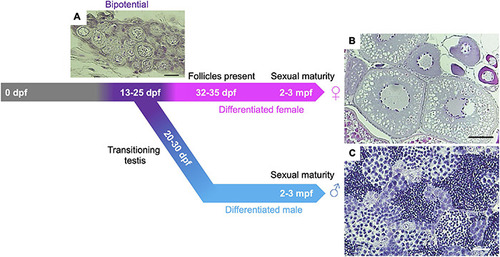
Gonadal sex differentiation and oocyte staging in zebrafish. Adapted from |
|
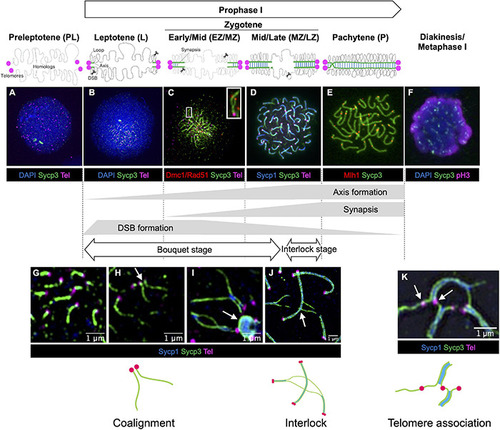
Stages of meiotic prophase I in zebrafish. Immunofluorescence staining of synaptonemal complex protein 3 (Sycp3) with telomeres (Tel), DNA (DAPI) and/or stage specific markers on zebrafish spermatocyte spreads observed by conventional immunofluorescence microscopy |
|
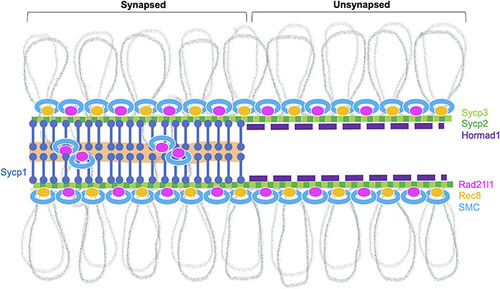
Meiotic chromosome axis structures. The synaptonemal complex comprises the lateral elements (LE, Sycp2 and Sycp3) that run along the lengths of homologous chromosomes joined by a central region that contains the transverse filament (Sycp1) and a central element (a region indicated in orange). Chromosomes joined by this tripartite structure are considered “synapsed.” Prior to synapsis, the LE is referred to as the axial elements (AE) where chromatin is organized into loops that are serially attached to the axis. The chromosome axis is made up of cohesins, the axial element proteins and HORMA-domain proteins. In zebrafish, axis localization has been observed for the cohesin components Smc3, Smc1β, and Rad21l1, the axial element proteins Sycp2 and Sycp3, and Hormad1. Localization of Rec8a/b (there are two paralogs in zebrafish) and Hormad2 (not shown) remains to be determined. Homologous chromosome pair at the ends as seen by the coalignment of axial elements. While the Rec8 cohesin complex most likely links sister chromatids together, it is not clear what DNA sequences are associated with Rad21l1 complex, however, it could play a role similar to the COH-3/4 cohesin complexes that enable the formation of asymmetric chromosome loops in |
|
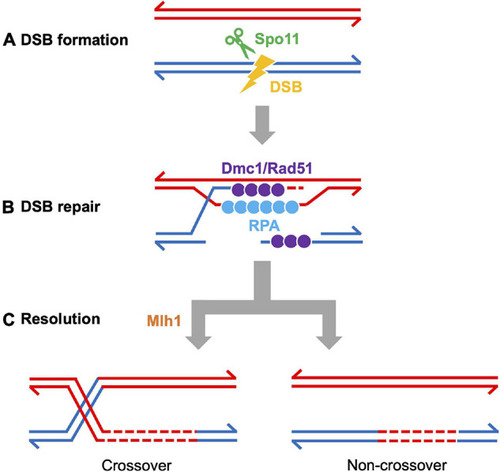
Meiotic recombination pathway showing steps mediated by proteins described in the text. Adapted from |