- Title
-
Usher syndrome Type 1-associated gene, pcdh15b, is required for photoreceptor structural integrity in zebrafish
- Authors
- Miles, A., Blair, C., Emili, A., Tropepe, V.
- Source
- Full text @ Dis. Model. Mech.
|
Pcdh15 is expressed in the photoreceptors of the zebrafish retina and is reduced in CRISPR-generated pcdh15b mutants. (A) Representative images of the 10 dpf retina stained for Pcdh15 (magenta). Co-label with F-actin stain, phalloidin (green; present in the CPs and synapse) is shown for the central ONL. (B) Close up of a central region in the ONL stained for Pcdh15 (magenta), phalloidin (green) and an overlap of the two. (C,D) Close up of the co-staining for Pcdh15 and phalloidin in the CPs (C) and for Pcdh15 at the IS-OS junction (D). (E) Representative images of SV2 staining (yellow) and calcium channel 1.4v staining (Cacna1fa; magenta) in the photoreceptor synapse and colocalization with Pcdh15 and phalloidin (green) at 10 dpf. (F) Schematic of Pcdh15 expression in the zebrafish photoreceptor cell. Green represents phalloidin in the synapse and CPs; magenta represents Pcdh15 in the CPs, OS base, IS membrane and synapse; blue dots represent calcium channel 1.4v in the synapse. (G) gRNA used to target and mutate the pcdh15b gene. A 17 bp insertion (magenta nucleotides) and a 7 bp deletion (black nucleotides) mutant were identified. (H) Predicted results of the mutations on the protein sequence. (I) RT-qPCR to analyse pcdh15a (left) and pcdh15b (right) mRNA expression, normalized to actin, in wild-type siblings and pcdh15b mutants. (J) Staining for Pcdh15 in wild-type siblings (+/+) and mutants (Δ7/Δ7 and ins17/ins17) at 10 dpf (n=5+ for each genotype). (K) Genotyping of clutches of heterozygous crosses up to adulthood to measure survival [numbers of fish genotyped (n) are indicated]. CD, variable cytoplasmic domain; CP, calyceal process; EC, extracellular cadherin domains; GCL, ganglion cell layer; INL, inner nuclear layer; IS, inner segment; ONL, outer nuclear layer; OS, outer segment; TD, transmembrane domain. |
|
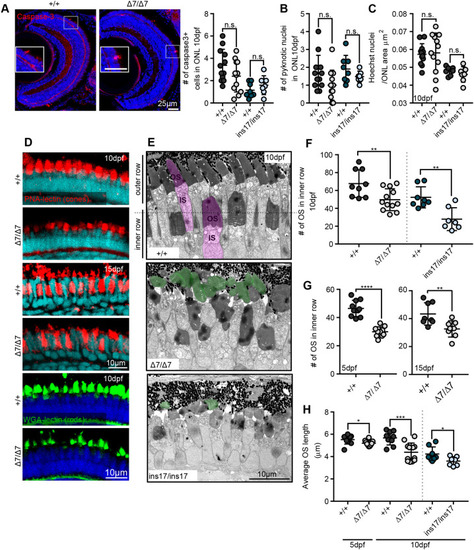
Cone and rod photoreceptors show reductions in OS number and length that are not attributed to cell death or loss. (A) Staining and quantification of the cell death marker, cleaved (activated) caspase-3, at 10 dpf in the ONL per section (+/+, 12 eyes) (Δ7, 12 eyes). (B) Quantification of the number of pyknotic nuclei in the ONL. (C) Photoreceptor nuclei (Hoechst) density/μm2 in the ONL at 10 dpf, measured from an average of 20 µm-thick sections. For B and C: +/+, 12 eyes; Δ7, 11 eyes; +/+, 8 eyes; ins17, 7 eyes. (D) Representative images of OS markers for cones (PNA lectin, red) and rods (WGA lectin, green) at 10 dpf and 15 dpf. (E) TEM images of the central retina of wild-type siblings (+/+) and Δ7 and ins17 mutants at 10 dpf. Magenta represents individual photoreceptor; green represents detached OS. (F,G) Quantification of the number of OSs in the inner row at 10 dpf (F), 5 dpf (G, left) and 15 dpf (G, right). (H) Quantification of the average length of all outer segments in the retina per section in wild-type siblings and Δ7 and ins17 mutants at 5 dpf and 10 dpf. Statistical analyses in A-C and F-H were performed using Student's t-tests (unpaired, two-tailed). n.s., not significant; *P≤0.05, **P≤0.01, ***P≤0.001, ****P≤0.0001. Number of eyes analysed for TEM data in E-G: 5 dpf: +/+, 10 eyes; Δ7/Δ7, 10 eyes; 10 dpf: +/+, 9 eyes; Δ7/Δ7, 12 eyes; +/+, 8 eyes; ins17/ins17, 7 eyes; 15 dpf: +/+, 8 eyes; Δ7/Δ7, 8 eyes. At least n=5 individuals were assessed for each immunostained marker. IS, inner segment; ONL, outer nuclear layer; OS, outer segment. PHENOTYPE:
|
|
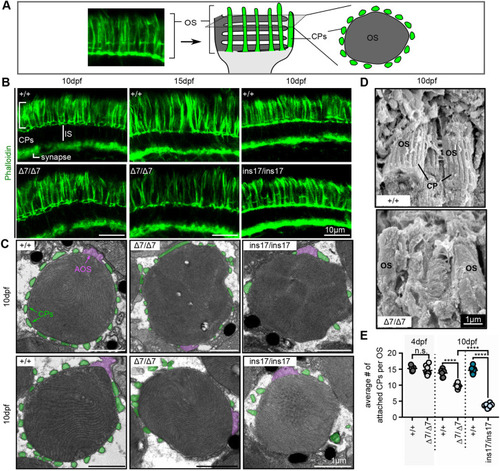
CPs are formed in pcdh15b mutants but are weakly attached and reduced/lost over time. (A) Schematic illustrating CPs surrounding the OS of photoreceptors. The F-actin stain, phalloidin, is used to label CPs. Transverse sections along the length of the photoreceptor show long CPs along their length, whereas horizontal cross-sections through the OS show OS surrounded by a ‘ring’ of CPs. In all images, CPs are coloured green. (B) Representative images of phalloidin staining in the photoreceptor layer in wild-type siblings (+/+) and Δ7 or ins17 mutants at 10 dpf and 15 dpf (n=8+ individuals for each genotype). (C) Representative TEM images of horizontal cross-sections through the OS in wild-type siblings and Δ7 and ins17 mutants. CPs (connected or disconnected to the OS) are coloured green; AOSs are coloured magenta. (D) Representative SEM images show CPs along the OS in wild-type siblings, which are absent/decreased in Δ7 mutants (5 eyes from n=5 individuals per genotype). (E) Quantification of the average number of connected CPs per OS in wild-type siblings and pcdh15b mutants at 4 dpf and 10 dpf. OSs quantified were sampled from the central retina in the ONL. Number of eyes analysed for CP counts: 4 dpf: +/+, 5 eyes; Δ7/Δ7, 8 eyes; 10 dpf: +/+, 6 eyes; Δ7/Δ7, 6 eyes; +/+, 8 eyes; ins17/ins17, 8 eyes. Statistical analyses in E were performed using Student's t-test (unpaired, two-tailed) for 4 dpf and one-way ANOVA for 10 dpf. n.s., not significant; ****P≤0.0001. AOS, accessory outer segment; CP, calyceal process; IS, inner segment; OS, outer segment. |
|
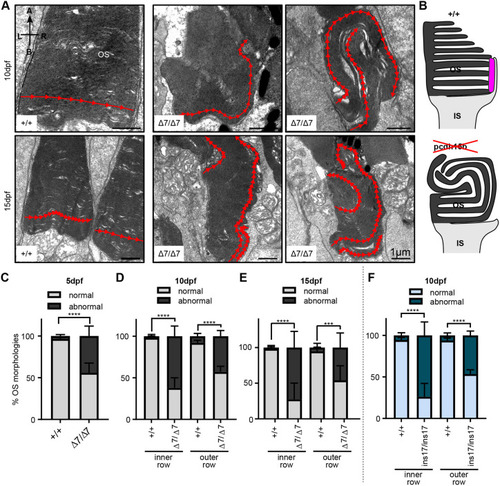
Ultrastructure of OSs in pcdh15b mutant photoreceptors exhibit abnormal directional growth of discs. (A) Representative TEM images of OSs of wild-type siblings and Δ7 mutants at 10 dpf and 15 dpf. Red arrow line shows disc direction. Directional arrows at the top left of the first image indicate positioning. (B) Schematic of the effects of pcdh15b mutation on the growth of the OS discs. Pcdh15 is shown in +/+ by the magenta-coloured shape. (C-F) Quantification of the percentage of ‘normal’ and abnormal OSs in wild-type siblings and pcdh15b mutants at different time points and mutant types (Δ7 and ins17). Statistical analyses in C-F were performed using Student's t-tests (unpaired, two-tailed). ***P≤0.001, ****P≤0.0001. OSs were analysed across the entire ONL for the 5 dpf and 10 dpf data, and the central retina for 15 dpf data. Number of eyes/OSs analysed: (B) 5 dpf: +/+, 10 eyes/2123 OSs; Δ7/Δ7, 10 eyes/1874 OSs; (C) 10 dpf: +/+, 9 eyes/1990 OSs; Δ7/Δ7, 12 eyes/2457 OSs; (D) 15 dpf: +/+, 6 eyes/320 OSs; Δ7/Δ7, 6 eyes/291 OSs; (E) 10 dpf: +/+, 8 eyes/1260 OSs; ins17/ins17, 7 eyes/956 OSs. A, apical; B, basal; IS, inner segment; L, left; ONL, outer nuclear layer; OS, outer segment; R, right. PHENOTYPE:
|
|
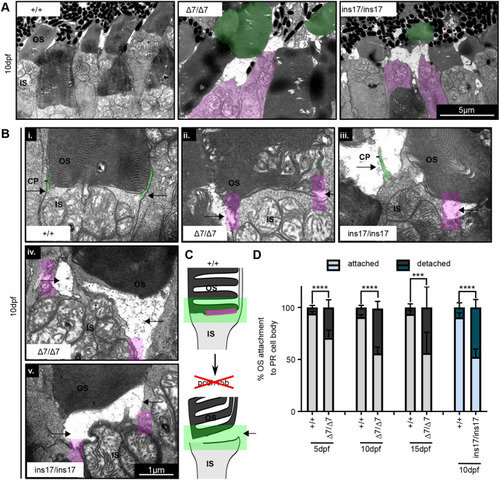
OS detachment from the photoreceptor cell body is increased in pcdh15b mutants. (A) Representative TEM images of OS detachment and floating OSs seen in pcdh15b mutants (Δ7 and ins17), compared to wild-type siblings. Magenta is used for IS with no OS; green is used for detached ‘floating’ OS. These phenomena are rarely observed in wild-type siblings. (B) Close-up TEM examples of IS-OS connection in photoreceptors of wild-type siblings and Δ7 and ins17 mutants, showing the process of detachment. CPs are visible as protrusions coming up from the IS (green) and attaching to the sides of the OS (arrows point to the sides). pcdh15b mutants (Δ7/Δ7 and ins17/ins17) show a range of affected connections (i-v), coincident with the loss of CP (magenta-coloured boxes) attachments on the sides (arrows). (C) Schematic of the OS detachment seen in pcdh15b mutants. The relevant Pcdh15 expression found at the OS base is highlighted in magenta. The green box highlights the IS-OS connection. (D) Quantification of the percentage of total identified photoreceptor IS/cell bodies with attached or completely detached/missing OSs across the entire ONL from 5 dpf to 15 dpf. Statistical analyses were performed using Student's t-test (unpaired, two-tailed). ***P≤0.001, ****P≤0.0001. Number of eyes/PRs analysed: 5 dpf: +/+, 10 eyes/1780 PRs; Δ7/Δ7, 10 eyes/1473 PRs; 10 dpf: +/+, 9 eyes/1844 PRs; Δ7/Δ7, 12 eyes/2290 PRs; 15 dpf: +/+, 6 eyes/334 PRs; Δ7/Δ7, 6 eyes/474 PRs; 10 dpf: +/+, 8 eyes/1253 PRs; ins17/ins17, 7 eyes/1087 PRs. CP, calyceal process; IS, inner segment; ONL, outer nuclear layer; OS, outer segment; PR, photoreceptor. PHENOTYPE:
|
|
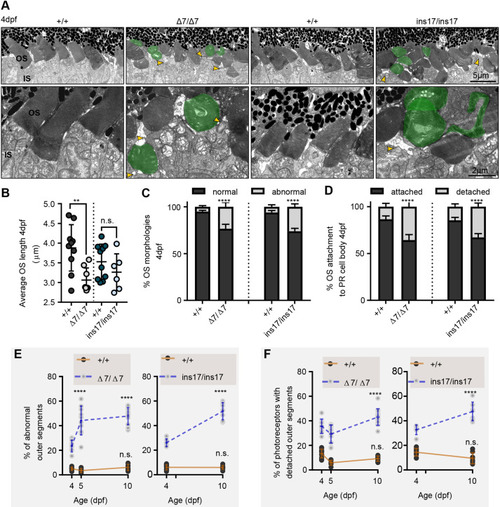
Photoreceptors in pcdh15b mutants develop with defects that progressively worsen with age. (A) Representative TEM images of the central ONL (top row) and OS (bottom row) in wild-type siblings, Δ7 and ins17 mutants at 4 dpf. Green represents deformed/abnormal OSs. Yellow arrowheads point to regions of disconnection between the IS and OS. (B-D) Quantification of the average length of all OSs in the retina per section (B), the proportion of OS morphologies (C) and the percentage of attached/detached OSs (D) in wild-type siblings, Δ7 and ins17 mutants at 4 dpf. The entire ONL was used for quantification. (E,F) Developmental trend of the percentage of abnormal OSs (E) and photoreceptors with detached OSs (F) in the retina from 4 dpf to 10 dpf in wild-type siblings, Δ7 or ins17 mutants. Statistical analyses were performed using Student's t-tests (unpaired, two-tailed) (B-D) and two-way ANOVA (E,F). n.s., not significant; **P≤0.01, ****P≤0.0001. Number of eyes/OSs analysed for 4 dpf data: +/+, 10 eyes/1528 OSs; Δ7/Δ7, 8 eyes/1412 OSs; +/+, 10 eyes/1829 OSs; ins17/ins17, 6 eyes/1328 OSs. IS, inner segment; ONL, outer nuclear layer; OS, outer segment. |
|
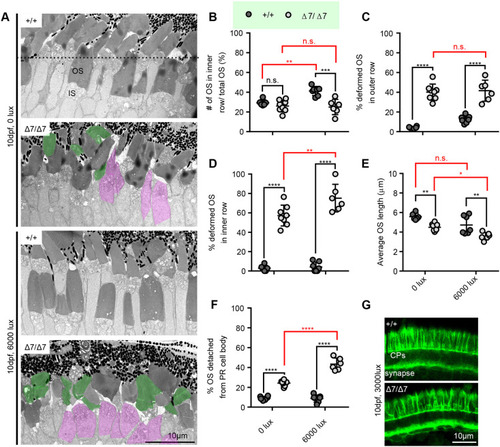
Differential light exposure in pcdh15b mutants attenuates or exacerbates photoreceptor defects. (A) Representative TEM images of wild-type sibling and pcdh15b mutant ONL at 10 dpf grown under 0 lux (dark) and 6000 lux (bright) light conditions. The dotted line in the top image separates the inner and outer row. ISs with detached OSs are coloured magenta. Deformed OSs are coloured green. (B-F) Quantification of the percentage of total OSs found in the inner row (B), the percentage of deformed OSs in the outer row (C) and the inner row (D), the average OS length (E), and the percentage of total identified photoreceptor IS/cell bodies with completely detached/missing OSs (F) across the retina photoreceptor layer under different light conditions in wild-type siblings and pcdh15b Δ7 mutants. All graphs in B-F use the same labelling as in B to distinguish between +/+ and Δ7/Δ7. x-axis labels are the same and seen at the bottom in E and F. (G) Representative phalloidin staining in the ONL, labelling CPs, at 10 dpf when exposed to 3000 lux bright light (+/+, 7 eyes; Δ7/Δ7, 7 eyes). 0 lux measurements were taken across the entire ONL, and 6000 lux measurements were taken only in the central ONL. Statistical analyses in B-D and F were performed using two-way ANOVA. Statistical tests in E were performed using one-way ANOVA. n.s., not significant; *P≤0.05, **P≤0.01, ***P≤0.001, ****P≤0.0001. Numbers of eyes/OSs analysed: 0 lux: +/+, 6 eyes/1239 OSs; Δ7/Δ7, 8 eyes/1623 OSs; 6000 lux: +/+, 7 eyes/541 OSs; Δ7/Δ7, 6 eyes/409 OSs. IS, inner segment; OS, outer segment. PHENOTYPE:
|
|
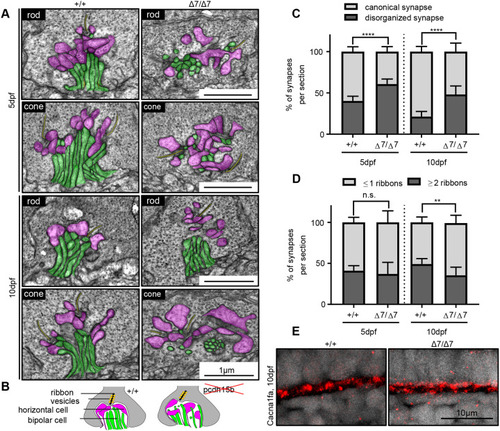
Photoreceptor ribbon synapses show increased disorganization in pcdh15b mutants. (A) Representative TEM images of photoreceptor ribbon synapses from both rod and cone terminals in wild-type siblings and pcdh15b Δ7 mutants at 5 dpf and 10 dpf. Bipolar cell innervations (green) and horizontal cell innervations (pink) into photoreceptor termini and ribbon synapses (yellow) are highlighted. (B) Schematic of synaptic structure in wild-type and pcdh15b mutants. Magenta, horizontal cell processes; green, bipolar cell processes. Ribbons are shown as a black line, surrounded by yellow-coloured vesicles. (C) Quantification of the percentage of synapses that display a canonical or disorganized synapse organization in wild-type siblings and Δ7 mutants at 5 dpf and 10 dpf. (D) Quantification of the percentage of synapses that contain one or less ribbons (usually associated with rod termini), or two or more ribbons (usually associated with cone termini), in wild-type siblings and Δ7 mutants at 5 dpf and 10 dpf. (E) Representative images of calcium channel 1.4v staining (Cacna1fa) in the outer plexiform layer, at photoreceptor termini (n=5+ individuals for each genotype). Statistical analyses in C and D were performed using Student’s t-tests (unpaired, two-tailed). n.s., not significant; **P≤0.01, ****P≤0.0001. Number of eyes/synapses analysed: 5 dpf: +/+, 10 eyes/620 synapses; Δ7/Δ7, 10 eyes/593 synapses; 10 dpf: +/+, 8 eyes/201 synapses; Δ7/Δ7, 8 eyes/192 synapses. Synapses used in quantification were sampled from the central ONL. |