- Title
-
The zebrafish presomitic mesoderm elongates through compaction-extension
- Authors
- Thomson, L., Muresan, L., Steventon, B.
- Source
- Full text @ Cells Dev
|
(A) |
|
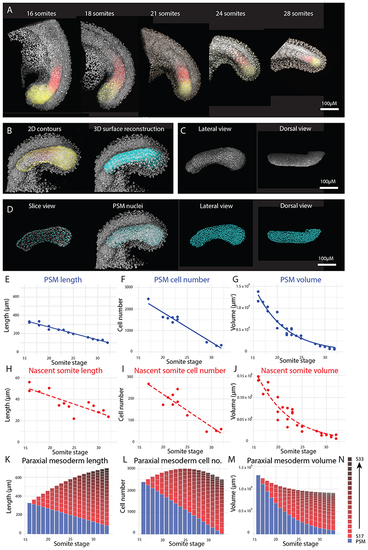
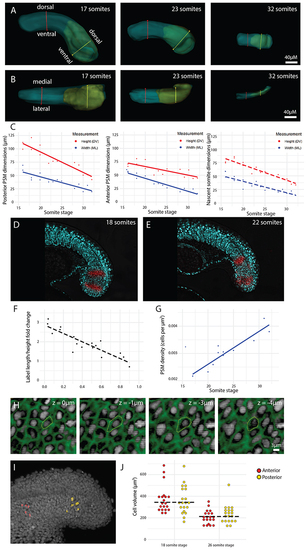
(A-B) Height (DV axis) (A) and width (ML axis) (B) measurements were taken of the posterior PSM (yellow line), anterior PSM (red line), and nascent somite (not shown) from the 16 somite-stage to the end of somitogenesis. (C) All height and width measurements show a decrease over the course of somitogenesis (n = 15 embryos). Solid trendlines indicate genuine change of one tissue (PSM) over time, whereas dotted trendlines indicate a trend based on separate tissues (nascent somites). The trendline equations are as follows (where |
|
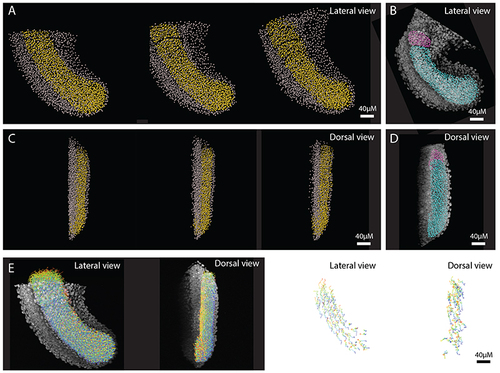
Tracks were generated of the whole tailbud and all non-paraxial mesoderm tracks were then manually removed. Paraxial mesoderm (PM) tracks (yellow spots) and non-paraxial mesoderm tracks (grey spots) shown at 0, 1, and 2 hours after imaging. (B) PM spots reconstructions (cyan: PSM & pink: nascent somite) from a similar-stage HCR image is shown for comparison/validation of selection accuracy. (C, D) The same images as above, but from a dorsal view: anterior is top, medial is left. (E) All PM tracks of full movie (colour-coded by time) superimposed over first frame image, shown for lateral and dorsal views (left images), and a sub-set of tracks shown in isolation, for lateral and dorsal views (right images). |
|
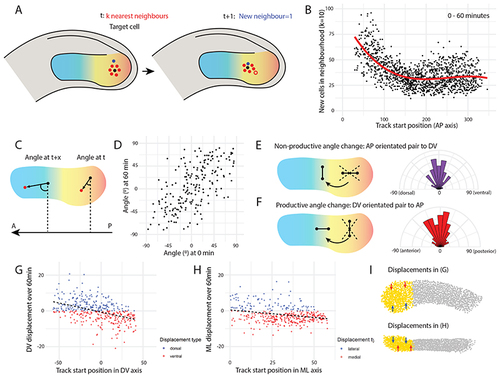
(A) Analysis of neighbour exchanges. For each paraxial mesoderm cell (black), a neighbourhood was specified as a given number (k) of nearest neighbours (red cells). After a given time interval, the number of new cells (blue) that entered the target cell’s neighbourhood was calculated. In this example, one new cell joins the neighbourhood (k = 6) over time, with the cell that is no longer in the neighbourhood shown in red outline. (B) The number of new cells entering each target cell’s neighbourhood (k = 10) over 60 min is plotted against the initial AP position of the target cell, with 0 being the posterior end of the tail, and ~ 300 being the anterior-most cells. Each point represents a single cell (n = 1,348 cells from one embryo). The results show that more cell mixing occurs in the posterior PSM (C) Analysis of neighbour angle changes. For each posterior PSM cell (black), the angle between the vector to the nearest neighbour (red) and the AP axis was calculated at the initial timepoint (t). After a given time interval (t + x), the same vector angle was calculated between the two cells (regardless of whether they were still neighbours). 0°: cells lie perpendicular to the AP axis. 90°/-90°: cells lie parallel to the AP axis. An angle change from 0° to 90°/-90° would indicate strong directional intercalation of neighbouring cells. (D) Measured neighbour angle changes are shown for 60 min. Each point represents a cell pair (n = 201 cell pairs from one embryo), with the x-axis value providing the initial vector angle and the y-axis value providing the vector angle after 60 min. (E-F) Comparing the distribution of non-productive vs productive angle changes. (E) Those neighbour pairs that initially lay parallel to the AP axis (DV angle > -45° & < 45°) were selected, and the distribution of DV angle |